lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01031359.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-460.40) is given below.
1. Sequence-
CAAAUUUCAUCUUUCAAAUAUUCAACUUAAAUAGAGGAAAAUUGCACUAGAGAAUUCUUC
UUUUAUCAAUGUGGGACAAUAUCCAAAUUUCAUUUUUCAAAUAUUCAACUCUUUCCUGGA
GUUAUAUCGUUAAUUUUUAAACAUUGACUUUAUAUUUAUAAUAUUAAAUAUAUUAUGACA
UAUUUAUAAUAUUAAAAUAUUUAUUACAUCAUUAUGACGUCAUCCGAGUAGAAGAGGUUU
UUACGUCCGUCUCAACCAGUCGGAGCUGUUGACCCCCGACCCCUAAGCUCAUCCGAGUCU
GAAAACAUUGCAUAUAAUGAACUCUAUAACAUAGCAAACAAGUAAUCAUUAGGCAUAACC
UAGAACAAAACCACAAUUAAAACAGAAAACAAGAAACAACCAAUGAACUCAAUUUAACAA
GUAACAUAUGCUCAGAUUCUGUUAGGCAGAAUUAAGAAUUAAUAGAACCAACUCUAUCUU
AGCUAUCAUAUCUUGACCAAGUCUUACAUGUAGAGUAGUUAGAGGGAGAUAUACAAGUAU
AUUACGAAAUUUUAAAAGCGCAAACAGAUAGGCAGCAUGAUAGUCAAAGGUUUACUGAGC
AGAUCAAAUUAACACUCAAGAAUAUAGCAGCUAUAUCUAGGCAGAGUAAAGCAGCUAACU
CAUAUGAACAAAAUACUAAGAUUAAGCUAAACUGAAUACAAACAGACAAGUUAAGACUAA
UGUUGGAUGUCAAUAAGCUAAUAGGCAACAGAGGGGUUACACAAGACCAAGUUAAGCUUU
UAUUAGAUAGUCCAUUAGUCAGCUAGUCAAGAAGAUGCAAGAAAGAAUCACACAAUGUUG
UUGAGGCAGGUGAUCAAACUGGAAAGCAAUAUAUACAAGUCUAUCAAUACUGGAGCGGGG
AGAUAUGCCUAGGGCCCAAUAAAGAGUACUCGGCAUUUUCAUUUACUUAAAAACAAUGGA
GAUCAUAUUUAAAUAUCAUUCAUCCCUCAAACCUUUGGACUGACUGGAGUUAAGUUGGAC
CCACACCAACUUGUGGUCCUCACCUUAGUAUUCUUGCAAAGUAUAAGCCACUAGGAUUGA
UAAAAAAAAGCCUUGGAUUCCCAGAGUUGACCUUACAAGUCUACUCCCUUUACUUGUGCU
CAUUAAAGUCCUUGGGCAAAUUAAGGGUCAUCUUUGAAAAAAACAGAAUGUGGGGUGGGU
UGAGGGGUUUUUGCUUUAUGACUUUGGGGUUUCAAAGUUAUUAUAUAAAAGUCACUUUGA
AGAAUAGGGAUCAAAUGGCACUAGGAUAGCCUCCCUUACCAUACCAAUCCUAUAGGCAAC
CUGAAUACUUUAGCCCUUACUUUUAAAAGAAAAAGAACUUUUAUAAAGGAAAAUAUUAGG
GCCAAACUAUGAGUGAAAUCUCCCCUUUAAUGAAAAGUAGGCAACAUUUAAUCAUAGACA
AAAACAAAAAACAUAUAGCAAAUAAGGGACACAUCUCAACUGAUAAAGACUAUUGGAAAG
CAUAAUAUCAUAGAGACAUCAAAACACAUCUUUUUAGCCUUAAAACCCACUCAAAAACAA
UACUAGCCUACAUAUUGAAGGUCAGACACAUAGCAUUUAGCAAACUAGUAAGUAAACUCA
UUCAACUACUAAGAAUCAUUAUCAUCAGAGAAAUAAGAACAUAACCUGGUUAUAAUCAGA
AAAAGGGACAACAUUUUGACUCUAGGCUAACAUAGAGCCAACAUUAUCUUUCAAUUCUAA
CUAAGUUCAAACAGUCAGUCCCAACAAACAAAGUCAGUUUCAUGUAACAAUUAAGAACCA
GAACCAGCCAUCAUGAUCAGAAAAAGACCAUGCUUUUAUUUAAGCUAUCACAUGGAAAUA
UUAACAACUAGACAAGUUCAGUCUAGUUCAUUCUAGGCAUGCAUAUUUACUUUUCAAUUA
UAUUAGAACACAAUCAAUGUAUCAAAAUGACCAGAAAUAGAAUCCUACUUACACAGGAAA
AUAGCAACACCAAAUCAAAUCUCAUAACGUCAAAACCACCUCCUGAACCCUCUAUAAAUC
AAAGUUAAAGCUAAAUCAAAUCAAGCUACACUAACAACUAAGUCCAUCCUGCCAUGAUGA
UAAGAAAAAGCAGCACACCAUAUUUUGAUUAUCAACACCUCAAGUAGAGUUAACUAAAAC
AGUAAUAAACAAAGCAGUAUCCAGCAGUGGCUAUAAACAUGCAAAAGAAUAGCUUAUGCC
AGCAAAUAAUGUAAUUGAUAGAGAAAAUAAAGGCCAAGGAGUAGUAUAGUACCUUCUUUA
GAGUAGGAACCAAAGUUGAACCAGAUAAAUAAUGCAGAAAGCUUAAAAUAAACUAAGAAC
ACACUCAAAGCUCUCUGAACCUACCAAACCUCUUGAAGGAGUGCAAGGAAAGCUUCUUUA
AAUGGAACACUCCAAGGUAGAGGAUCACUCUGCUUUUACCUAUCUUAGAACUGAUGUAAG
CUUAAAUUGUGAAAUUAAAGAGCUAAGUGAAAGCACCGCAGUAAGGUUCCAUCCUAGAAA
CACCUUACUGCACCAUUAUUAU
2. MFE structure-
....................((((((((.......((....((((.((((((((.((((((....((....(((........))).....................((((((.....)))))).....((((((........))))))..((((((((......))))))))..(((((....(((((((..........
..........))))))).)))))..))..))))))((((((.(((((((.((.....)).))).((((((................((((....))))(((......(((.(((((.(((......(((.((((.....((((((.......((((..(((((((.......................(((.(((((...
...................((((.....))))..((((((((...))))))))........(((((......)))))..((((((((((((...(((...)))....)))).)).))))))(((((((((((..((((((............(((((((...(((.((((..((((((..(((((..(((((...(((..
...)))....))).))........((((.(((...(((....)))....))).))))...........................((((...(((........)))...))))..)))))))))))..)))).....(((....)))........((((......))))..)))..)))))))...(((..((((((.((.
....(((.((((((((((..((.(((........((((((((....(((.........)))...)))))))).......))).)).....(((.(((((........)))...)))))..............)))))))).)).))).....)))))))).)))))))))..))))).....))))))........((((
((((((.....(((((((.......)))))))(((((......(((.(((....))).))).....)))))(((((((((.............(((....)))((((.(((.......))).))))((((((....((((((..........))))))...))))))...(((((((..........(((((((((((((
(..((((((((((((.((((..((((...(((((((((.(((.......))).))))))))).((((((.....((((....(((........)))..)))).....))))))(((...)))..........(((((..((((((((((........)))))).)))).........)))))....((((((.((((...
..((((((.....))))..)).....)))).)))))).......))))...)))).))))))...((((....))))..((((((.....))))))...............(((((............)))))..))))))..)))))))))..........((.((((.((...)).)))).)).))))).((.....)
)....(((((.((...........))))))).............)))))))..............(((((....)))))....((((.(((....))).)))).((((.......))))...........)))))))))............))))))))))...............))))).)))...............
.......((((..((((..............((((......)))).)))).))))..........((((((((.......))))))))..))))))).))))....)))))).....)))).)))........))).))))))))......)))........((((........)))).))))))...............
......)))).))))))..(((((...((((((((.............((((...........))))..(((........))).....((((..((.((((((..((((.............))))..)))))).)).(((.....)))...(((.....)))..........))))......(((..((((((......
........))))))..)))..(((.....))).....))))))........))...)))))............)))))))).))))...)).)))))))).......(((((((((((.(((((.......................))))).))))...............((((((((((.........)))))))))
).((((....))))((((((((.....)))))))).....((((........))))..(((((.((((....))))...)))))..(((....))).((((((((((..............))))))))))..)))))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-506.02] kcal/mol.
2. The frequency of mfe structure in ensemble 7.12466e-33.
3. The ensemble diversity 610.18.
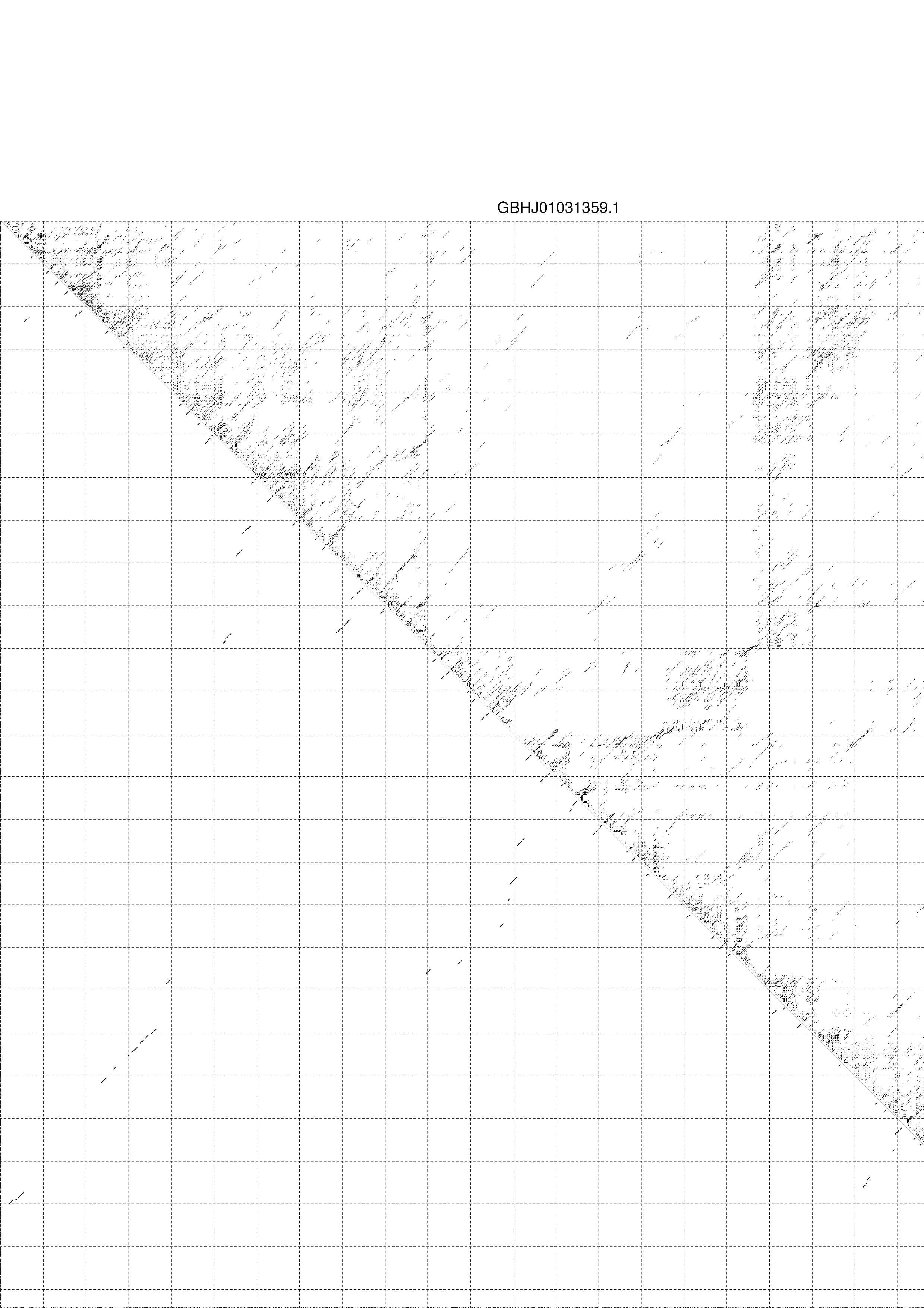
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
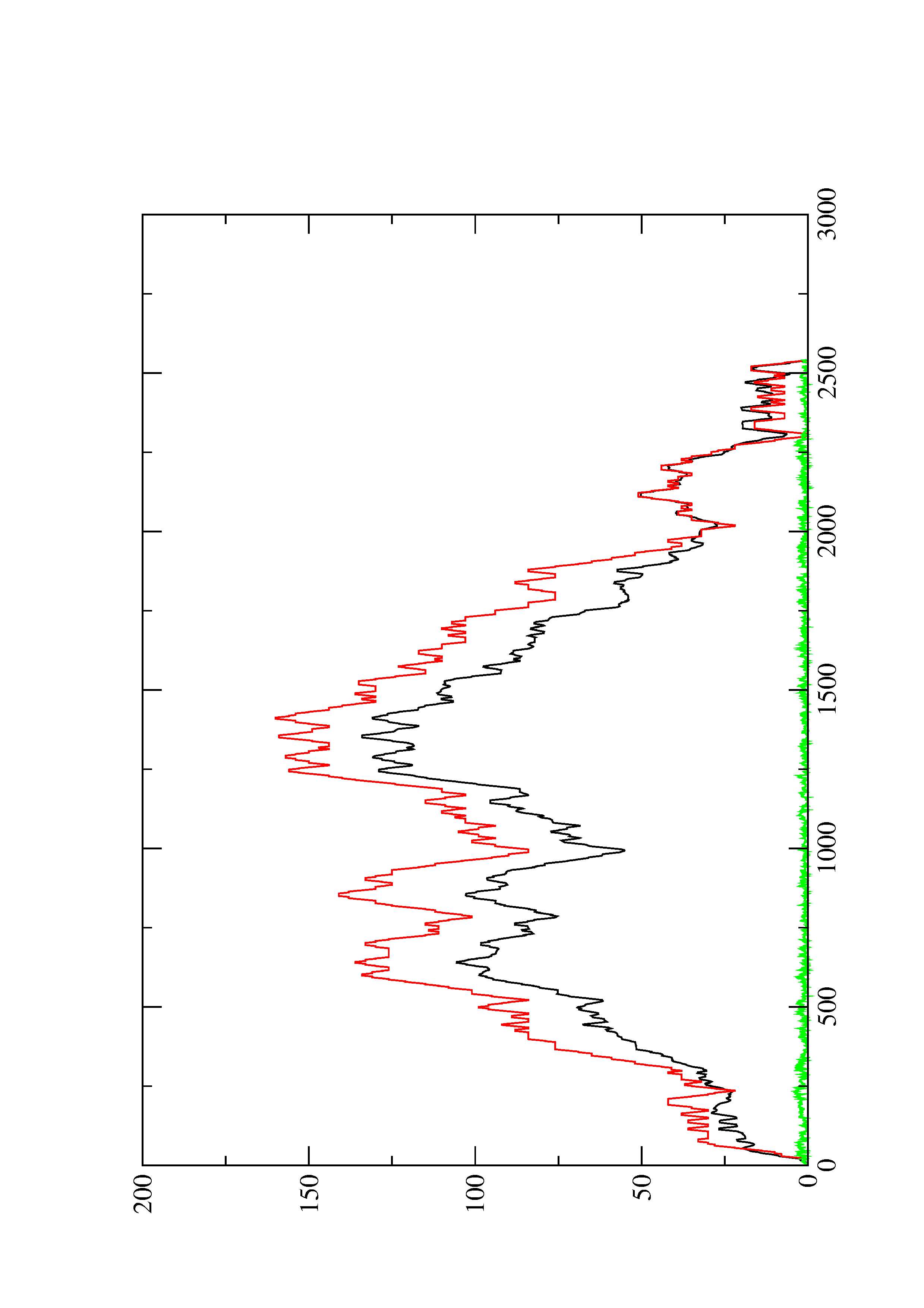
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.