lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01033350.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-492.90) is given below.
1. Sequence-
AUCUUAUAUAAUUUUAAGUGUUUUAGAAUUUUAGAAUUUAAAGGUUUGACUUCAGUCAAC
UUUUAAUAAUUGGAGCCUCGAAUUAAAAUUUUGAUUGCAUUAUCAACUUCAAAAUAUAAA
UUUCUGGUUAGUUAAGUGGUUUUUUGUUUCCGAAGCUUAUUUUGAAUACUCGGUUGGAAU
UACAAAAACUUAAGUUGAGGGUUGAAUUUAUUUAAUAUUUUCUAAAACUAUCAUGGAUCA
AAAUUUCAUUUGCUUCGUUGAGUACAAAAUGUCAUUUUCAUGCCGUUAACAAAUUUGAUU
UGUGUUCAUCGGGUUGUAAACUCAUUCUGAACCAUCGUUUGAAGUUUGAAAACUUGGUUG
AAUAUUUGUCAUCUGGCGUCAGGCUAUUGUUCUUCACGAUCGCGAGUCCAUCUCACAUGA
UUGCAGUGCACUAGCAUUUUCUUCUUCACGAUUGUGACGUUUAGUCACGUAAUCGCAUAG
UUUUGUUUUCCUAUUCUUCACCGAUCAACAUGUGGCUUCACGCGAUCGUGAUUCUAAGUC
ACGCCACAGUGUAGAGUAACCCCUAAAACACUAUAAAUAUCUCAAAACCCAAAUCUUCAA
CUUGUUUCUCCAUCUCUCCACUUAAAAAUCCAAAAUUCUCCAUAGAUAAACCUUGGGGUU
UUGCAUGUGACCUCAGGAACGUUGGGAGUCAUAUUUGUUCAACUUUUCAUCAAUUAGUUA
UAAGUUUCCAUUUUAUUCAUCAAAUUUCAUUCAAAAUAUCCCAAAUUUCACCAAGUUCUU
UCUUCACUUUAAUUCUUUGAUAUUGGCUCUUUCCUUGCUCAUCUCCAUUCCAAAUUUAUG
UUUAAGUACUUUGCGAUGAGCGAAAUAUUUUGAUGUCUUUUGUUUGAAGCUUCGAUUCAC
UAGGCCCAUCGUCCUGUUUUGACGCGGAUUUUGACCUUUUCUAUAAUCAUAGAUUUAUGG
GUAUUGUUUGUCUCUUAUUAAUUGUAGAUUAUUAUUUUGCUAGUUUAGUAUGUGUUAAAG
AUUUUUUGCGAAGAUGUUGGAGCUGUCUAAUCUAUCUUUUUUAUAUUUGACAGUUUGUUC
UUCGUAUGACAGGUAGGUAACUGAUCUGAAAUUGGGUUUAUACUUUAUAUCAUAAUAGUA
AUUAGCUAUCCUUGGGUUGGAUUUUUAUUAGGUUUGGAACCCUUAACCUUGUCCGCUUCU
UGGCUUAUGAUUCGGUGUUGCAUUAUGAUUGUAGAUUACCUUGUGUGGUUUUUAAUCGAC
UUUGGAACAUAGAUUAUGAGAUUAGCGAGAAAUAUUUGUGAGUCCCAAAUAAUAUGACUG
GUUAUAAUUGUUGUAGUGACACAAUCCUAUGCAGGGCCCACAUGACUUGUUAUAAUCUAU
UUUUUAAGGUCCAUGCAUGAAAUGAUCAUGAAAAUUUAUUUUUAUGCAUUGGUUUCUACU
AUUUUAUAGACAAGGGCCCGACGUUACUAAUUUUGAAACCUUGUACAGUUGUGUGGACUC
CUUUUUAGUAUUAUGAGAUUAAAAUGAGUAUAAGAUGAUAAUUUUACCAGUUUCUAAUGA
UAAAUGAUUAAAGAAGGAAUUUCCCAUUGUUUUUGAAAUAAUAUUAAAGUCGGUUAAGAU
AUGGCGCUAGAUAUUGAGAUUUGUACUAAUUUUAACUCAGUAAGGGCCAAGAAAUGACAU
UGAGCUUAUAAUGAUUUAGACUCUGUGAAGGCUGAGAACUUAUUUUGAGAUUAAUAUUUU
GAGAUAGACCUUGUGGAUCCAUGCUAUGAGUUAUUGAGAUGGUAUGUGGUGAUGGCCUCC
AUGGACCAUUCUGAUCUGAGCUAGUGAGAAAUCAGAGGAAGAAUAUACAGAGGAGUAGAG
GUAUACGGGUGAAGGAUUUUCAUGGAUUCUACCUGUUGUACCUACGGGUAUUGGAUGAGG
AGCUUACAUGUUUUUAGUAGAAUCAUAUCAUAUCAUUUUUCAUAUUGUACUCUUGUCUUC
UAUCUUUUGGUUGUACUCACGUUAUAUGGGUUGAUGGGGAGUUUGUAUUCACUUAUCUGU
AUCCCUUGGGUGUAUGGGCGCUUAUUGGGAUUUUACUGUAUUUAUUUCAUAUAUUCGUUU
AGUGCUAUAUUGAUAAUCUCUUUCUUCGAGGUUUAAAAAAAAA
2. MFE structure-
.(((.....((((((((.....))))))))..)))..(((((((((.(((....))))))))))))......((((((((((.....(((((((..............)))))))...(((((.(((....((((((((.(((((((((((((....((((...)))).....))))))..)))))))..((((((((((
((((((..(((...)))..)))..........((((((.(((((..(((((((((....)))))...))))..))))).)).)))).........((((..((((((((((((((.((((((...((.((((....))))))))))))..))))))).)))))))..))))..((((((..((((........(((((((
((((((.((((......((((((.(((((....)))))..........((((((((((.....)))))..)))))..........................))))))....))))))...))))))))))).....)))))))))).(((((..((......))...)))))...................)))))))))
)))).............))))))))....))))))))..(((.((......)))))((((........))))(((((((((.((((((....((((.((((((((................))))).........((((.((((...(((.((((((((...........((((..((....................))
..))))........(((((((((..((......(((((....)))))....)).))))))))).)))))))))))...))))..))))((.((((..((...(((.....))).))..)))).))........((((..(((((....))))).....))))((((((((((..(((..(((((.(((((((((((....
(((((...(((.(((((.........((((((((.....((((((((.(((.((.......)).))).))))))))..))))))))))))).)))...)))))((((......))))((((((((....((((((((.(((..((..((((.......)))).......(((((.((....)).)))))..((((((...
...(((((((((..(((((.((...((((((.(((((((.....))))))).))))))....)).))))).)))))))))...)))).)).........((((((......(((((((((.((((((.....((((.(((.((((..(((((((((((.............................))))).)))))).
......((((((((....)))))))).))))))).))))....))))))((((((...(((.(((...))).)))...)))))).)))))))))))))))))..))).)))))))).........)))))))))))))))))))...)))))..))).))))))))))....))).)))))))))).)))))))))....
.....((((..(((((.(((((((((((((((((((((((..((((.........(((((((((.(((.........(((.(((.(((........))).))).)))..((((.(((..((((((((((((....)))))))))))).((....))(((((((((((...........)))))))).)))(((((((...
..)).)))))....))).)))).(((((.((((((((((((((...(((..((((((.((((((((((((..(((((((....)))))))))))).)))))))...(((((..((((..((..((((.........))))..)).))))))))).............))))))))).)))).))))))))))...)))))
..((((((((..((((((((((....)))).)))))).....)))....)))))..))).)))))))))........))))..)))))).)))))....))))))))))))..))))).)))).)))))))))).........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-534.79] kcal/mol.
2. The frequency of mfe structure in ensemble 3.01918e-30.
3. The ensemble diversity 527.72.
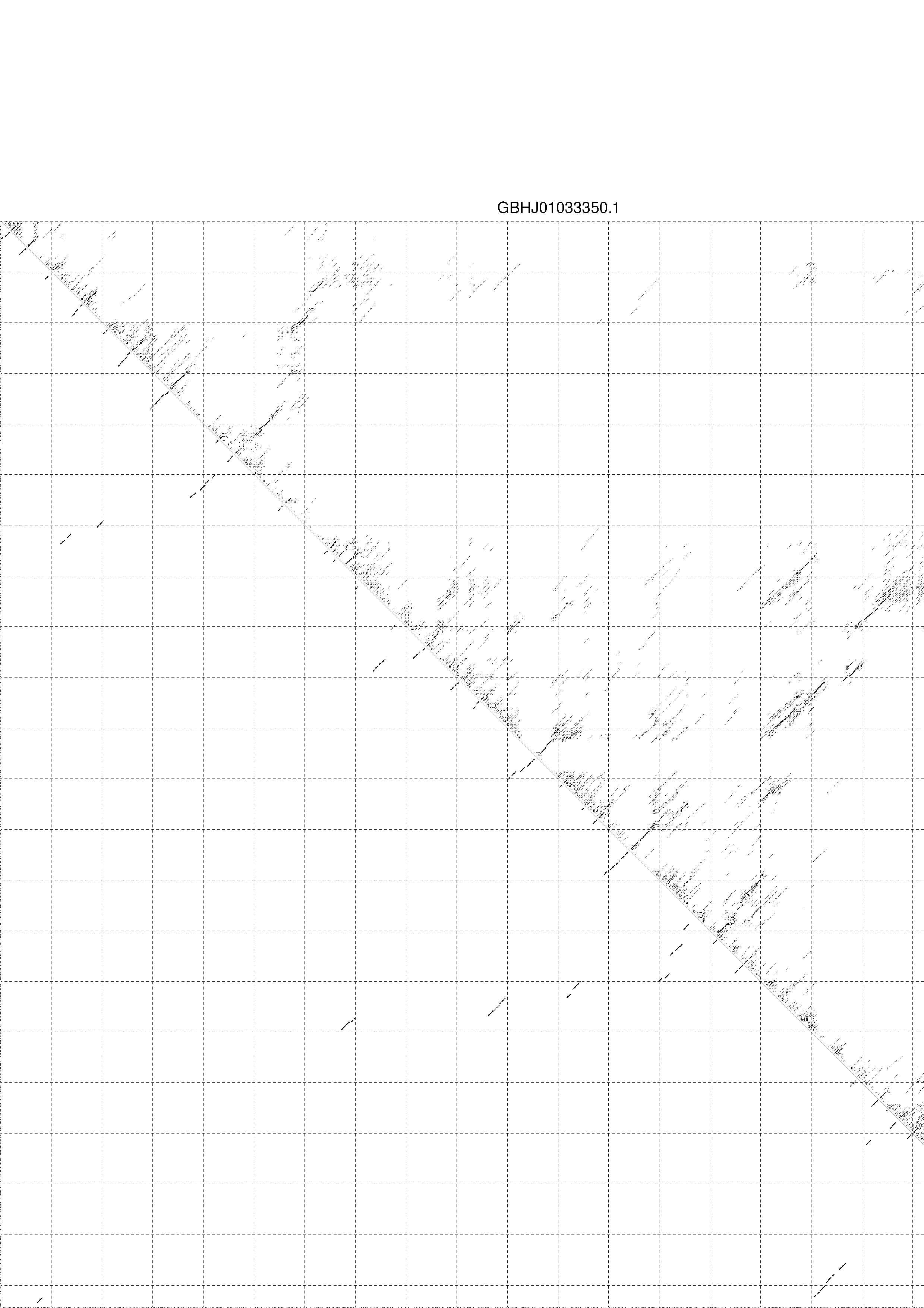
4. You may look at the dot plot containing the base pair probabilities [below]
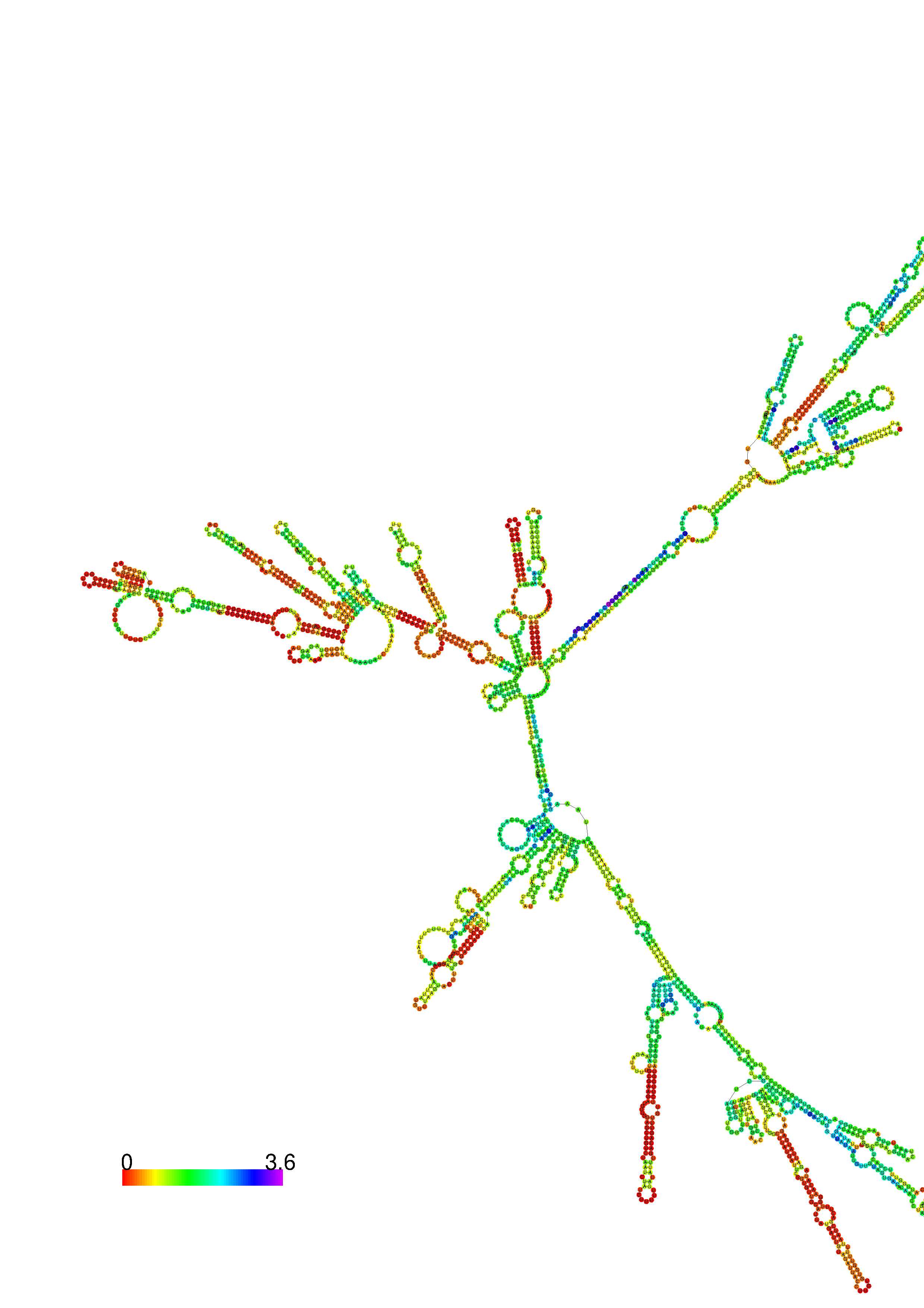
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
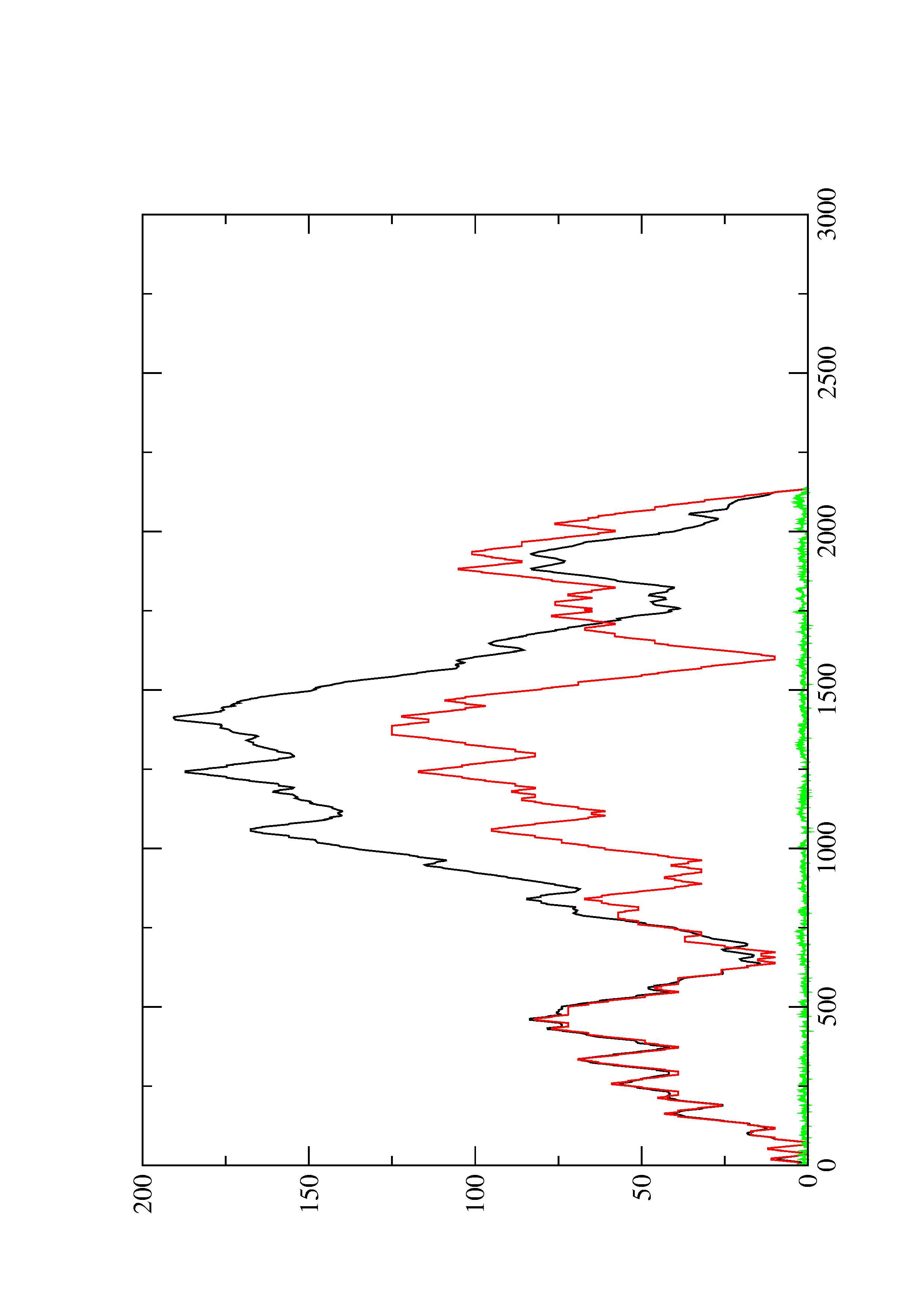
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.