lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01033814.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-331.94) is given below.
1. Sequence-
AGUUGAAUCACGAGCGAGGGCCUGUACUCUGAUCUUAGUCCCAUUUAGUUAGUUUUUCUC
UAAUCUUCACGCCUUAUGGUCUUCAAGUAUUUAGCUCACCCAAGGUCUUGGGUUAAGGAU
UUGACUCGAGGUUCAUGCAAGAAAGAUGAUAAAAUUAGCACUUAUCAAUGAGUCCAAACU
UUUAUAUGAGUAAAAAUCAAUAAUGCCAUAAUGCAUAAAACCUACAACAAUAGCAUUAUC
UCAUGAUAAAAAAAUAUGAAAAAAGAUACAAAAUACCAAAGAGGAUAUCACAUGGUAGCA
UACACGCUAAAAAUAUACAUGAAAAGUAAGGUUGACAAAGAAAAUCUUCCCCCUGCCUUU
CCCCUCGAAAACAAUUAAGAGAACAGUCUCAAAGUUCUAUUAAUAAAUCCUACUCUGUAU
UAUGAAUCAAGAGAAUUUACAAGAAUAAUCCAUAAGUCUACAACCAAUACAUCAUCUUUA
UAUGACACAGAAAAAAUUAUCCACAACAAAUGUAACACUAGAUAUCCCACAGAUAACAAU
UAGUGAAGGUUUCAAAAAAAAAAUAUAUCGAACAAUAUGCAAGUAUUAUACAGAAUGGUA
AGCCCAUAUGGAGAAACCCAAAAAUCACAUUCCAAGAGUAUACUGGACCAUGUUAGGAUA
GUAAAAAAAUCUUUACAAUUUGAUUACGUCUGUUCCACUAGAAAAUUUUUGUCAACGAAU
GAUUACAACAGAUUAUUUUUUUGUAAGAAAAUCUCCUUCGUACCCCAAACUCAAACCAAA
CAGAGUCCUACACAAAAAUUUAACAUAAUGAAAAGCUUUUCAACAUGAACACCAUUAUCG
UUCAUUAAAAUUAAAAAGAACCUGAACAUACCACCAUUUAUGAAUCAAUAUCACAUCACC
UAUUUUUGUUCAAAACAUACCUUUUAUAAGCUAAAUAUCCACUCCUCUUAUCACUUAUAU
UAUUUUAUCAUGUCAAUCCACAGAAAGUAUAUCGAUCUUUAGCUAUUCAAAUGUUUAUAC
UAUUCAUAAUCAAAAAUCAUAAUCUAUCAAACAUAAUUUAGCAUACAAACAUACCCUACA
GAAUAGUCUAUUCUACCAACAUCUCACGAUUUAUUUUAUAAAGAAUGAACGAAGAAGGAA
UCACACUUUAGCAUGGAGCAAUGGAAGAUAGAAGCAAAUCAUGGGUUGGGUAGAAAUAAU
AUUUCCCACGAUUCUUUGUAACAACUUUUCAUGAAUAGAGACAAGUUAUGUAGUACCCCC
ACAUUAAUAUUCUAUGAAAUUUUAAGCAUACAUGCGCAUUUACUAAAACACAAUUUAAGG
CUUACAACACUCUUAUAAUCCUGAGCAUGUGAAUCCUAAAGCUCCAUAAUUUAUUGGAUG
GCACCGACUGACUUAGACCGAUACUAUACAGUCAAAUGUAAAAUUUUUCAUAUUUUACCG
UAUUAAGAAAUAACCAAGACAAGUACAUUCAAGUGAGACAGGAUUCUAGAAGGCUAACAC
AUGUAGUAAAGUACAGAUCCCAAACAUAUUGAUGCUCAUGGAAUCUACAUUAGAUAAGUC
AAGAUAAACAUGCAAACCAAAAUUUAUACAACUUAGCUAUAAGAAAGAUUCAUUCUUGAA
UAUAAAAUUUUGAAGACAUGAGGUUGCUUAGACCUAAACACAUUGUUACACAUAGUUUAA
CAUCAAUCAACUAUUUCACCCAAAGGUAUAUUCAUAGUAAUAACACAACUUAGUACACUA
UUUUCCAGUAGCAGAUUCAAGACAUGAGAGAAAAUACAUAUUUGGAUAAAACCUAUUUUA
UUUAAGUCGGACAUAGUUUGGAAGAAUUAAAUUUUACUUGUUCAAGAGUGAAACAACUAC
UUAUCCUGCCACAAAAAUAGUUUAACCUCUGGAUUUAGACCAAUGAUAAAGGGAGUUUCC
AUGUUCAUUUUGUUCUUCUAAUGACAACAAAGGUGAACAAUGUCCAGACCAGCAAAAUUA
ACCUUGAUUAAACACUUAUUUUCAUAUUAGGUUCAAUUCUUUUAACAAGUACAAAUUCUA
CAACAAGAUAUCUUUACACAUGCAGCAGAAUCCCACAAACUAUCAUCAAUCAUACUAAAU
AUCUAGAAAAGAUCAAUACUGGUCAUGUAACCAGGAAAUAUUUAAAUAAAAAGUA
2. MFE structure-
.(((........)))(((........)))(((((((.(((.(((((.((((((((((((((..((((((.(((((..((((...((((.(((((((((((...)))...)))))))).)))))))).)))))...)).)))).))).)).)))))))))..((((((.((((.....((((......)))).........
.......(((((((..................))))))))))))))))).(((((((((((((...((((...........(((((.(((((((.((((......))))........((((((((((...((((.((((((..(((((((.......((((.....))))...(((((.(((((.........))))).)
))))..((((((.(((((.(((((....((((..........................................))))..))))).)))))...........(((.....)))..((((((.((((.....))))....))))))..((((((....................(((((....)))))...((..((((..
...)))).))..))))))........((((((((.........))))..)))))))))).(((((......)))))..............(((((((.....((((((((((..(((((.((((((((.(((....))).)))))).......)).))))).......((((..........)))).....)))))))))
).......(((((....)))))..((((((.........)))))).................((((((............(((.......))).............)))))).................((((((.......(((((((.......................(((.((.....((((((.((((......
.((((.....(((((((((.........................))).))))))....)))).................((((((...))))))............)))).)))))).....)).)))...))).)))).......)))))).)))))))..)))))))........(((.((((..(((((.......)
)))))))).))))))))))))).)))))))))).((((((.......((((.........)))).....))))))..........(((....)))((.((((.............))))))........(((..........)))))))))))))))...((((((........))))..))...(((((...(((....
...)))..)))))...))))...)))))))))))))...(((((.................)))))....))))).))).(((((((.....(((....(((((.((((((.....(((((((((.((((....((((((((((((....(((.(((((....(((((..............)))))...))))).))).
.....))))))..((((((((...................(((((.....)))))........((((((..((((((...........))))))...(((....)))......(((((....((........))..)))))......)))))).))))))))))))))....))))...((((((((((((....)))))
)))))))........)))))).((((((......((((((((.(((((((((............((((.....((((((((((((..(((((((.................(((...))).(((((((((((((..((....)).)))))).)))))))..)))))))....(((........))).))))))...))))
)).....))))))).))))))..))))))))..)))))).......)))..))))))))))).)))))))))).....................(((......)))..)))))))...(((((......))))).....................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-373.14] kcal/mol.
2. The frequency of mfe structure in ensemble 9.29629e-30.
3. The ensemble diversity 500.68.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
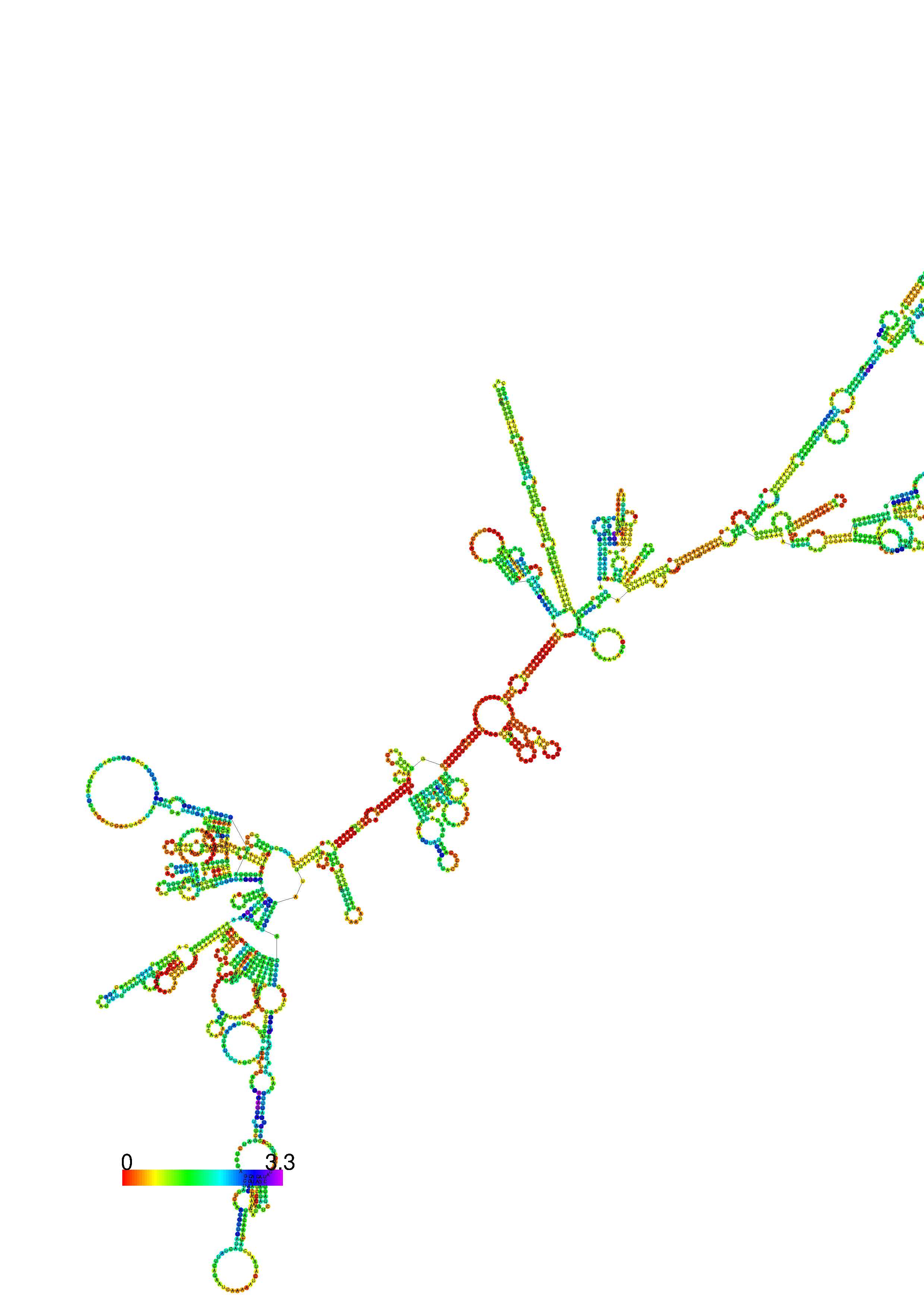
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
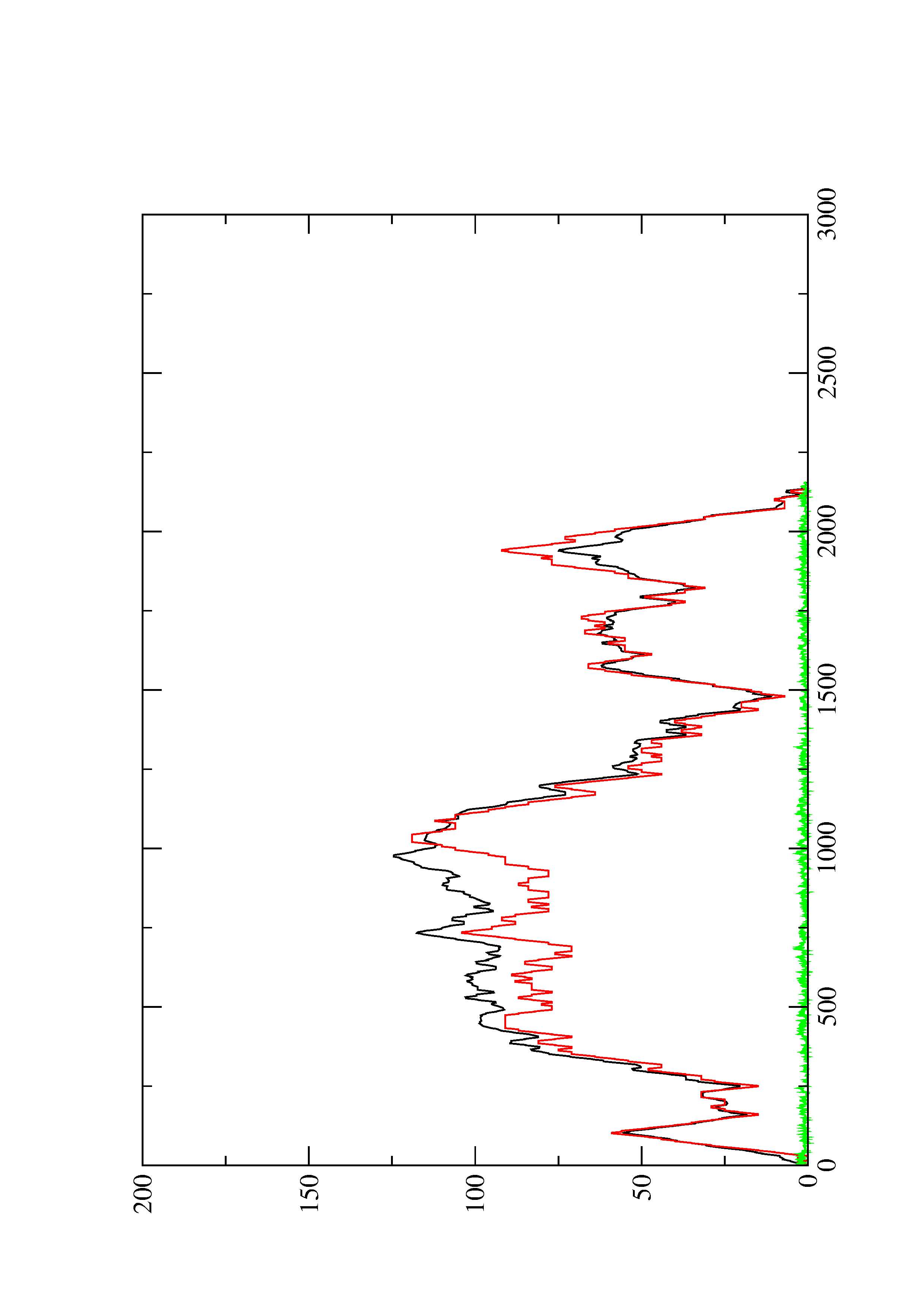
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.