lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01034560.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-666.40) is given below.
1. Sequence-
UAACUGGGUCAUAAAUUUUGAGGAUACCUCAAAAUUUCCAUCUGUAUUACCGACGUUCAG
AGUUGACUUAAAGUAUCCCCAGAUUCGAUAAGGUUAAGGAAUCACCUCAAUAAAUCCCCC
AACUAUGAUCCGACAUAUCUUCAGAAGUUUCCAGAGACUGUUGAACACAACAACACAGAA
GACAACAAGAAGAAAUAGAUAGUUAGAGGAAAUCAGUGUAGAUCAGUUUCAAAGGAAACU
AACAAUUUUUCUGUGAAUGUAGGUUUUUCGCGAAGAAAGGCCCAAGAAAUCCAAUAAGAA
GCCUAAAAGAGCUAAUACCAAUGGUUAACAGGAUGUAUUCACUCGCUUAUCCGGAAUGAG
CUACUGAGUUGGAGUAUAGGCGCGAAGGCAGAGAGUUACAUUGUUCACUGCUUUGUAUCU
AAAACAAUAUGCAGUUUCCCUUCAGCCUUAUUACGCGGGGAAGUUAAUUCCUAAUUUUGU
CUCACAUCAUGAGAUAAGUACGGAGGGAUAAGGUUAGUUCUUGGUUAUUGUUUUCUUAUU
UAUUUACUUUGCUCAUAUAUAUAUAUUUGCUAAUUGAAUUUUUCUGGGACUCUGAGCUGA
GACAAAGGUCCUCAAUACUUUGCCUUAACUCUUGAAAGUCCUUUGUUUUUUCUUCCUUAA
UAAGACCAGCCUUUAUAUGUAAGAUAUAGGAUAAUCUUGAGUAUGCAUUUAGCAGUAUUG
UUCUUUGUUCUUUUGAAAAAUAGUUGACGGUCAGACUUGUUCUUGAUCAGAACUCCCUUA
ACCUGCGACAAAAAUGAAAGUUUUAUACGUGCUAUCUUCACGAUAAAAUUUAUGAUUCUU
UGUUACAAUUAUGGUUCUAAUUGAUACUAUCUACUAUAUGGAAGAUAGAAUAUGCUGCCU
GGGACUUGUCUACCAUGUGAUAAUGUGAUUAAAAUUGAGGCCAGCGUGCAUGCUAAGUAG
AUUAAGGUUUGGUUUCAGAGAUUUGUGUGCAUGUUUUCCAUCUCUUCAUUUUCUUGUGUG
UUUAGAAAGAAAGUUUGAUUCCUUAUGUAAUCUCACAUUGAUAUCUAUUUUUCUUGAGAC
UUGCAUUCCCUGGUAAAGUCUUCUAUUCAUGUCCUUUGUCUGUGUUUCAGUCUUUUAAGU
GUCGUGUCAGGAAAUAAAUGCUAGCAUUGGUUUUUCUUGGUUUUGAGUGGAGUUGAUCAU
UCCAUGUCUCUUGCUCUUAGAAAUUAUAAUCACUUAGAUUUAGGAGGAUUAAAACUCGAA
CCUGUGUUGUGUCAUAGUAUGAAGUAGCAUGGUCUAAGUGUCCUUAGUUUUAAGGUCAUG
GUUGUGUCUAUUGAGUAAGUAAUGGAUCAACCAUGAUAAUUUGGUUCUGUAUGUUGGUUA
GUUUAGGUUUCCUCAUGCACCUAGCCUGGCAAUAUCUAAGUCAUUUUGAAUGUAGUUAUU
UCCUGCUCAUCGUGGUUAAGAGUCCAAGCGUGCCUGUUUAGUUAGCCGGGUUGAUCUAUA
UAUCUAUUUGCUGACGUUUCUCGAGUUCAACGAUCAAAUGAGUUAUGGUAAAUGUUCAGC
UACUACCUGAGGAGUAUACAGUGCUUACUUGGAAAAUAGACUCUCCUUAGGCUCAAAAAC
CAGUGUUGGCAGCAUGUCACUUAUCUCCUUAUUCUCUUUGACCAAUUCUUUGUGUGUUUC
UUGAUGUCCUUUGAGUGGUGGAAUCAUGUAAUCUACCUCGGGUAAAUAUCAAAUCUUUAU
GACAUGAUUUCCUUUGUUUUCUUUCUUAUUUAAAAAAAAAAAAAAAAAAGAUAAAUGACU
GUUUUUGUUUGCUGUUCUUGGAUCUUCUGUCCGAAAUAGAGCUAGCAAUGUUUUCAUGCU
AAAAGUAGAAGAACUCUCACCUUUCGUGACUAGUGAUGUAAGUUAUGUCGAUGACUAUCU
UGGAGUUUCUAGUCAUAGUUCACUAGUUGAAGGCAUGCCAAUAUUUGUCUUUCAUGUUGG
CUGGUGUUGCUUGCUGAUUUUGAGUAUAUAUAAGCAUAAGUCACGUGGGAAUAAAGCUAC
UUAAAGGCUGUUUUUGUUGGAUUCUUUUACUCUGUGUCUAGCUGCUUUAAAGAGGGAAUC
CCUCACAGUUUAUUAUUUUUUAGCUGGUCUAAAUGAGUUUCAUGCAUUGUUGCUCUUGUU
UGUUGUGAGUAUAUAGGUCAGCAGGUUUGACUGGAGAAUGGGUCACCUGGUGGUCUGUCU
UAUUACGUGCAUACCUCUAUAUUCUUACAAGUUUGCUUAGUUUUAUACAAUAACACACUU
GUUAAAUACUCAUCAUUUUCUCUUUAUUGCAUGAACUUCCUUUCUGAGCCCAUUGGACUC
AUCUUCAUUAUGGGCCCAAACCACAACAUCAAGCCAGUCCUCAAUUCCAUGUCCCGUAUA
GACCGAAGAUGGCAAGGAGAAGAAAUAAAUGGACCAAAUCCAUUAAGAGCAACAACAUCU
GAGGGCCCCAAAGCCCAUUAUUUUGUUCAUUUUAAUUUCAUUUGUGGGUUGGCCCUAAUA
UAUUUUCCAUACUUUAUUAUUUUUAUAUUAGGGGCCGAAAGCCUAGUAUUUCUAAAAUUA
GGACAGGUUGAUUGAAAUGGGCCGAAACCCAUGUAUAUCGGGUCCAGUUCUGUCACAGGC
GCAUAGAUAAAGAGAACCGGGUCCAAGUGGCUUCAAUCGAUUCCAAGUCAAGACAGUUCU
CUAUAUCCUUGAUUAUUCUUUGAUUAUUCUUUGC
2. MFE structure-
...(((((....(((((((((((...))))))))))).............((((.......))))............))))).........(((((((((((..................((((..(((((((.((((..(((((.(((...((((((((..(((((((((((.(((((((((((((((((((...(((.
.....(((((((...(((((.((.((((((.((((((...(((((.......(((((((..((((((((.....)))))))).((((((..(((((....(((...((((((((((..(((((..((((....(((.(((...(((((((((((....((...))...)))))...)))))).))).)))....))))))
)))...(((((.((((.......))))...))))).(((((((.((........)).(((((.....)))))...(((((((((.....)))))))))...)))))))....)))))))))))))))))).))))))...)))))))...((..((((....))))..))...............((((((..(((.(((
(.(((((..........))))).)))).))).....))))))))))).....))))))....((((((((((((((((.....))))))..((((((..(((((((((...((((..(((......(((((.((((.((..((((........))))..)).))))..))))).....))).))))(((((((((..(((
((((((.((((.......)))))))))))))...))).)))))).((((((......))))))...((((((.((....)).)))))).....(((((((.(((((((((........))))).))........)).))).)))))))))))))....))))))))).)))))))..)))))).)).)))))..))))).
)))))...))))))))).)))))...(((((((...(((.....((((......))))....)))...)))))))(((((((((........)).)))))))....)).)))...)))).)))))))))))))))((((((.(((((...........(((((((((((((((((...(((.....((((((((((.(((
((((((.(((..((((..((((((......((((((((.....(((........)))....((((((((..((((...))))..)))))))))))))))).(((((....)))))(((((((((.((((((((......)))))))))))))))))......((((.((((((..((.............)).)))))).
..)))).((((...(((.((((((..(((..((((.......)))))))..)))))).))).........))))......((((((.((((.(((......))).)))))))))))))))).)))).............))).)))))..)))).)))))).))))((((((((.........((.....))........
..))))))))))).)))))))))))))))))))))).))))))........(((((((((.(((..((((((.......((((((((((((((((((((((((((((((...........(((....)))...........))))))))))...................................(((((((......)
))))))..(((((((((((((((.....)))))....))))).)))))((((..((((((..((.(((..(((((..........((((((((.(((.((((.((.(((...))))).))))..))).))))))))))))).))).))...))))))..))))(((((.((((((((.((((.(((((...((((.....
..))))......))))).)))))))))))))))))))))))))))))........(((((((.............)))))))(((..((((((((((((......(((((...........)))))......((((((.....(((....)))...(((.(((.(((((...((((((((.(((((..((((........
))))))))).))))))))))))).))).)))...............((((((.((.(((...........))))).)))))).....)))))).))))))))))))..))).(((......)))((.((((((((((......))))..)))))).)).......)))))))).................(((.......
)))))))))))))))))))))......((((((.....)))))).........))).)))))((((......))))....(((.(((((((((((((..((((((...((((((((((((((.....................))))))).)))))))...((((((.........)))))))))))).)))))))))))
)).)))......)))).))))))).))))(((...)))...((..((((.(((((((...(((....(((..((....))..)))......))).))))))).))))..))...))))))))))).........
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-718.76] kcal/mol.
2. The frequency of mfe structure in ensemble 1.28048e-37.
3. The ensemble diversity 722.39.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
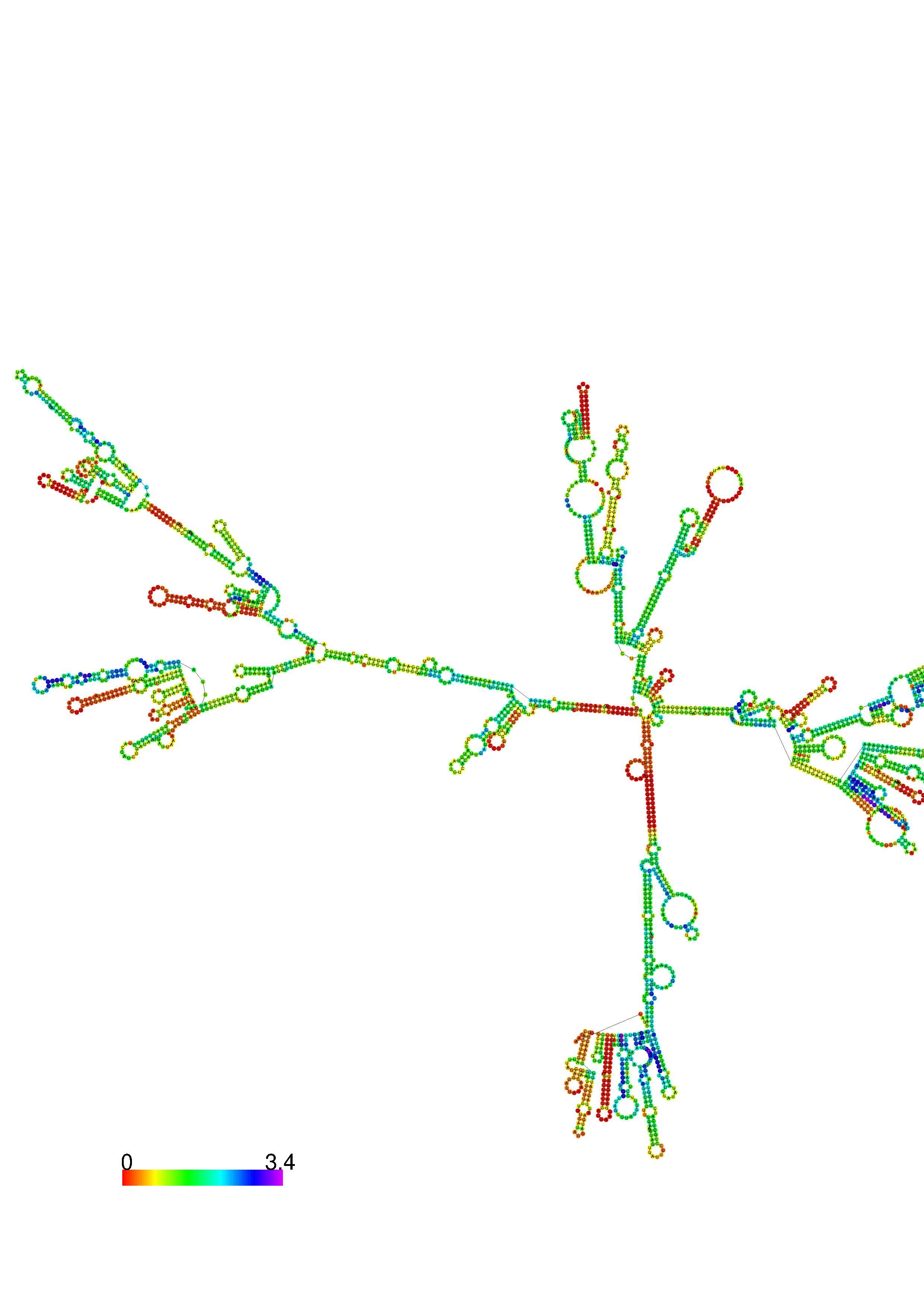
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
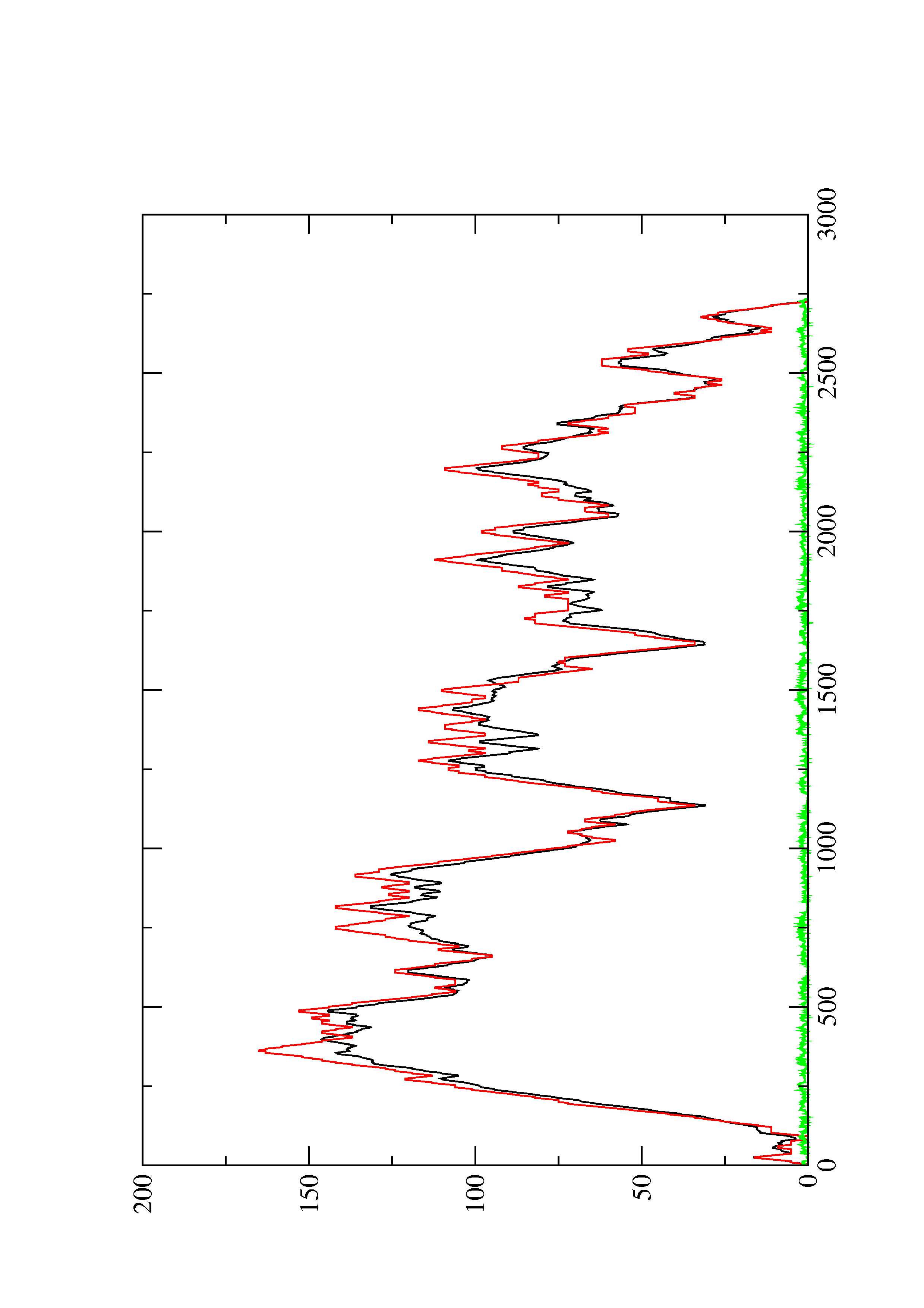
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.