lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01035823.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-545.10) is given below.
1. Sequence-
UAAUGAUGUAUAGUAGAGAAAAUAGCCUUUAAUUAGUUGAAUGCUCUCUGAGAACCUUCA
UUCUGCCCUGCAUACCCCUUCUUAAGAAUAUGAAAAUGAUUAGUUUAUCUAAGAGUCUUU
CUUGAUGUAGAACUUAGUUCUAAAAUGUCCGUAGAUAGGUUUUGAAUGCUGAAAAUGCUA
GAGAUUAUCGGGCUUUUGAACUUAUUUAUUGUGUGCCUUUUUAUGCCUGCACUGCUUGCU
UUGUUUUUCUUUUUACGAAAUGUGUUAGCCCAAAUGACUCAGUGAAUGUCUUCUAUUUGA
AUUCCUACUACAACACAUUUAUUUCCUGAUUAAGAGCACUGGAAGUAUAAGUCUAUAAAG
AUUGAUCGGCAUGUUCUACUUAGUUGUUAUUUGGUUUCCCAGUGGUGUCAUUUGAAAAAG
UGAAGGUUAUGAUUAAAUUGGUUUAUGAUCACGAUUUAGUUAAAAUUUGAUUUUAUGUCU
AAAGGGUAACUAUGGAUAGUAUGGGAUGUCUCAAAGGUUGUCAUGGUUGCGUUAAUAGAC
UUAGGUUCCUUAUCUAGGAACAUAAAUGUUGUUCCUCUUUCCUUUUCGUUGUUAGUUUUG
CAUUCUGCCUACUUAGUUGUUGUUUUACUGGUGUUUAUAGUAGUUGAUAUUUGGAUUUGC
UUGGGUUUUCUAGUUGUACAGCUCUAUACUUUAACUUGUAUGCAGCUAAGACCAUUAAUU
UUGUACGCUUACAAGACAAUUAAUUAGAUUUGACAAAGUAAAUAGGAAGUCUAAUUGCCC
CGCUAGAAUCAGUUAAUGAUUUGAAAUCCUUGGUUGUUUCUCUGUGCCAGUUUUAUUACC
AUAGCAAAUCAACAUGAUAUUGGUUAAGACAACCAAUUAUGUUGGAUCGUUGUAGCUAUG
CUCUACUCCAUGAUAUUAAAAUCUAUUUGGAUACUGAGUUCCUGUUGAUGUUUUUUUAUA
UCUAUCAGUUUAAAGUCUAUCACUUAUGCUAGAAAAUAAAUAAUAUAUGAUACAACAAAG
AUUGAGUUUUACAUUUUUAUAAUCUGUCUAUGAUUUUGCAUUUCUGUGAAGUUCCUAUUC
AAAAUAUAGGUGAAAGGUUCCUUUAUCUCCUCAAGGUCUAUCCAAUUUGUAGCCUUCUAA
AUGCUAGAACAUGCUCAUCUGUAUUGUAAUCAUGAAACUUCCCAUUUCACAUGCUUAUAA
UAUGUGAGGUAAAGAUCCUGUUUGGAUCAAUUUUAUUGCUAAAAAAUCUGAAAUAUGUUU
AUACAUGAUAUGUUUUAGAAUCUCUACAUAGUCUACAGUCAUUUAGAUGAAUCUAAAAAU
GGAGGGGAACCAGUUCUAUUUUAUGCUAUUUGACCCCUGUUUGCAUGUAUUAGCGUAGUC
GAGUGAAGUACAACUCUUUGUGUGUGCUUGUAUGGUCGUUAAAUGUGUUUGAAAGCUUAU
AGAGUGUGAUGUGUUGCCUGUAUUAUGACAAUUCCAUUACAACUUUGUAGAAGUUUCCAA
CAUGCAUAAAACCUGUGUUUAAUGUCUAGGGAUUACCUAGAUUACUUGAUACCUGCUAAA
UAGACUUAGAAUAUUUGAUAACUUAUUCGAACAAAGCAUGGAAUUGAUGAGAAACUGGAA
UGGAUAACAUAUGAAGUGGUAAAUUUGUUCUAUGAAACUGUCUAACCAGUUGUUGGCAAA
UUGCUUUGCAAAGAGCUAACUGUUUUUGUGUGUGGGCUUCCUUGAACCAUGUAAAUUUGA
AAUAGGAUUAUCCAUUUCAGUUAGUCAUUAGUGUAUGUAAAGUAUAUCCUUUGUUUGAAA
AUCCAUGUGUUUGUCUUCUAAUUAAGGUUCUCGACUUCUCUGUUAUGAUCGCGUUUUGUA
CCCCUACUAAGAAACCGAGCCGCCCAGGCUGCAAGAGACAAAGAGGCAGAUUCUCAGACU
GAAGAAGCAGUGGCUACCAUUGCUGAAGUUUGUCCAAUUUUCAAAAUGAAGAUAUUUUGA
UUUUCCUCCAAACAUGUUGCAGAACGAUGACGAAAGUAAUUGAUAUUGAUAGAGUUUGCU
UCCAACACUCAAUUAGUAGGACCCCCCGUCAGUCCACUCCUUCCCCACACUACGAGUGAA
AGAGAAAGUAAAUAUUACCAUUGAUUAACAUAUUGGAUAUAUAUUAAUUCUUUUCUUCUU
UUCAAGCAUGAACCUCCCCUCGAGUCAUAUUGGGACUCAUCCACAAUUUUUGUUGAGUCA
ACUAAAUGGAAUUAGGAAUUGAAGACCUUCAAUAGAAAAAAAAAAUUGGGCCCGAGGCCG
GAUUCCCAGGAUCAUCCCUUGAACCCUUACUUUUAUUUAUUUUGUUUUGUAUUCUUAUUU
GACUUGCUUUGUAUCUAAUACUAACAAAUUGUAAAAUGCAGUUUAAAAUGAAUUCCAAAG
2. MFE structure-
.((((.(((.((((((.(((.......(((((.(((..((....((.(((((.....(((((.((..(((.((((...(((.((((((...((((((((..(((.((((.((((.....))))))))((((((...))))))........((((((((((((.(((..((((.(((.......))).))))..))).)))
))))))))).((((((........))..)))).)))....)))))))).)))))).)))..)))))))..)))))))))))).))...))..))).))))))))))))))..)))))))........(((..((((((..((....(((((..((((((((((((.(((...((((((((.(((((....((((...((.
...))..((((((....))))))(((((.(((((((((((...........))))))))))).)))))((.((((.(((((.((.((((((((((......((((....)))).......)))))))))).))..))))).)))).)).......(((((((.......)))))))........(((((((((.((((((
((.(((((((.(((.(((.((((....((((.((.(((((.((((((((.(((((...((....))..))))).))).))))))))))......((((((.((...(((((.......)))))...)).))))))....(((((((((...((((((...((((((((.((.(((((...(((.((((.(((((((.(((
(...)))).))))))).))))..))).))))).......((((((.((((((((((((..(((((.....))))))))))))))....)))...))))))......((((.((((........))))))))....)).))))))))(((((......)))))(((((((((.((((((((....((((((((....((((
((.((((.((((.(((..((((.((.................)).))))..(((((((((....)))))))))(((((.......))))).....(((((((....((((...(((.((((.......)))))))(((((((((((((..((((((((..(((((((((.((((...............)))).))))))
)))))))).....(((((.....)))))..................(((((((((((((.......))))))))))))).......)))..)))..)).)))))))).............))))))))))).))).)))).)))).))))))....)))...)))))..((((((..(((((((((((((((((.....)
).)))))))...............(((((.((((..(((((((((((((((.((((............))))..)))))).)))))))))...)))).)))))(((((.....))))).....(((((((.....)))))))))))))))...)))))))))))))).))))...))))).....(((((((((((((((
..((((((((..((((((((((((((((((.....)))...((((((((......(((((......)))))...))))))))(((((....)))))...(((((((..(((((((...........))))))).....)))))))..)))))))))))))))..)))))))).))))...........))))))))))).
.........)))))).)))))))))((..((((..((((..((.......((.....))......))..))))..))))))))))))...))))..))).))))))))))....((((((.....((((((((...))))))))..))))))........((((((((....)))))))).................)))
))))))))))))))(((((................))))).))))....)))))))))))))...)))))))))(((((..............))))).((((((((((.((((((((((.............))).)).))))).))).)))))))....((((..((((..(((.....))))))).))))((((((.
..((((...))))))))))...(((((((((.((((.((((((....))))))..........((((.((((...)))).))))....((.(((.....))).)))))).)))))))))......))))))..)))))....))))))))..)))..........((((((((...))))))))................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-590.88] kcal/mol.
2. The frequency of mfe structure in ensemble 5.46501e-33.
3. The ensemble diversity 544.52.
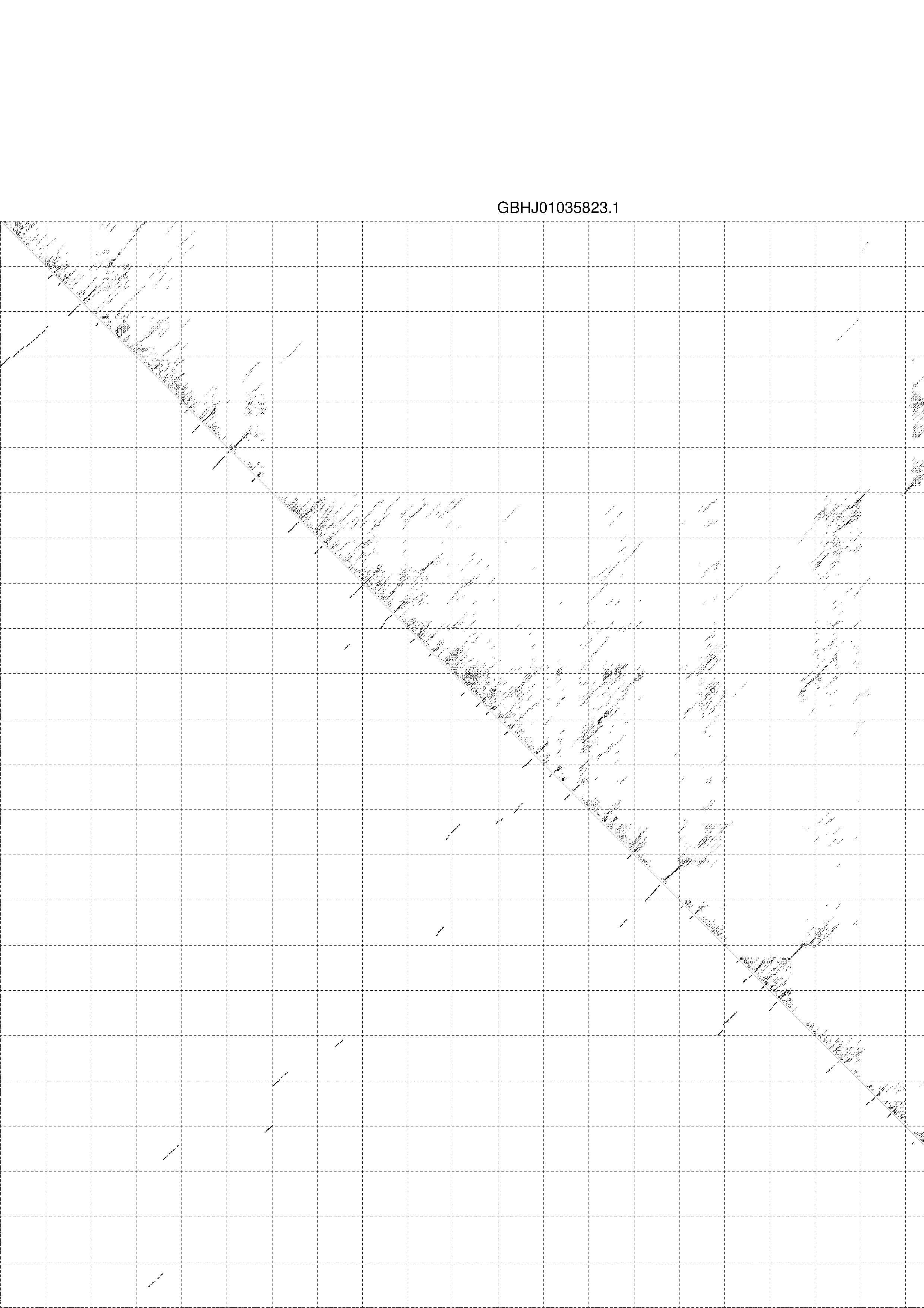
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
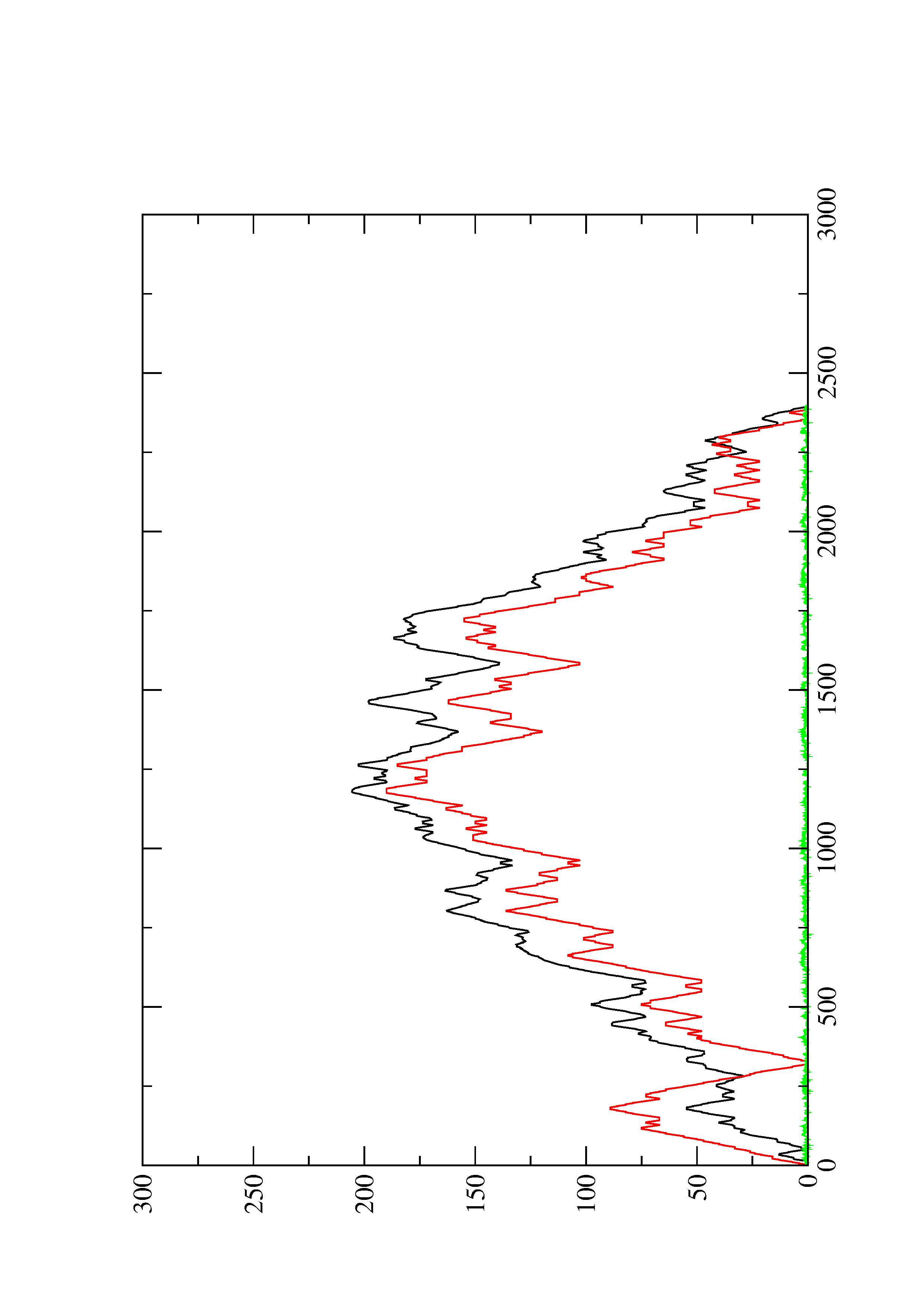
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.