lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01036035.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-431.70) is given below.
1. Sequence-
CUUUUUUUUUUUUUGUUAGUCAGAAUUUUUUAAGACACUUGAGUAGCUCACAGUCUUACU
GAAGUUCAUAGAGUAAACAUCCUUUCCCCUGUUCCUCUUUAAUCUCUUUUAAACUUUAUG
GUGGACGUUCUUUUGUUCGAGUUCAAGUUCUAUGGCAUUACAUUUUAUUUGAUCUGCGGA
UGAAAAUAUCAUCUUUGGUUUCUAGAAAGUUUCACUUUACUUCUUGAAACAUGAUUUAUA
UACUUUAAUUUGGUUCAAAAAAUCCCAUUUCAUGAUUGUUGGAACUAGUCUGGGAAUGUU
AGGACAGUUUGUUGUUGUCUAAAUGAGUGUGUGCUGGGUCAGUGAUCAUUUCAUAUUGAA
UUUCCUUCACCUUGUCGUUUGGAAUAUGUUUUAAGUAUUCUGUUGUCUAAAAAUGAGUUU
AACUUUCACUCCAGUUUCCUUAACUGGUUCUGUCUUUCUUGCUUUUGUUGUGGAUCCGUU
GCUUACUUAAAGCUGGUUUAGGUAGGUCUGUUGCUCAGCCCUUCACAGCAUGUUGUGGUG
CUUUGAUUGGAGUGUGUUAGUUCCUUUAGUCACCUGGGGAAGGGCGGUUAUGGGUUCAAA
UCUGGCUUUAUUACUUGAGCAUGGCAUUUGAAAACCUUUUCUCAUCUUCCUAAGUUUAUA
CAACCAAAUUCUUACUUCCAACUUUACGACUUCCUGUAAGGCUAUUCACAACCAGUCCCU
UGCUUUUUUUUUCCCGCUUUUUAGCUUGAUGUUUACCUAGAGUUGAUCUCUUAUACCCUC
UUAGUUGUUGUUUAUCUUAAUUUGAUCUUUGAUUCUACUGUUCAAGAGAUGCUGGCUUUG
UUUCUCGUCAAUUUCAUAUUUUGGUAGGUAUUCUCGUCAUGUAAGCUGAUAUGAUAAUAC
AUGUCUAGUGAAUAGGAGAAUGUUUAACCAUUCAUGUUGCCCUAUUUCUAGUCAGAACCU
GUUAUUGUUUUACUGGUUGUUGAUCUUGUCAAAGCUGAAGUUCCUAGGUCUUAUUUCUAA
ACUAACCUAGUAACAUGAUUAAGGAAGCUUAAAACUGAAACCUUCAUGUUUAAGCCCUGU
UGUAAAAGCUAGCUCUUGAAAUUUGCUCAUCCUUCGUGUUUCAAAUUUGUGAAAAAAUCU
UCAAAAUGAUGACCCCUUCAUACUGGAACUGGUGUUUCCUUUUGAAGUACGAAAUGUUCU
UGAGUUCAAGUUUUAGUUUGGUUAAAUUCUCGUGUACUUCCAGUGGUUAGUCACUAGGUC
UUUGUCUGAAUUAACCUAAAAAGGGCUUAGCUUAAUGUCACUUGUAAUGCCUGAUUUCUA
CAUGUCACCUGUUUAUGUGUCAUUUUCUUGUUGUCUGAGUAGUGUGAUCAUCUGGUUUAA
GACAAUUACUUUUCUCAGCAUAGCUGGUUUGGUGCCUUUCUUAACCUGUGAGUGUGAAAC
UUCGUGUGGAUUUCAGGCUGAGUAAGUAUGGUCCAUUAAAUACUAAAUGUUGUUUGUGCU
UUGUCGGUCUUGUUGUUUUACUUGUUCAAAUUCCAGUCUUGAUAUUACUGGAUAGGUAAC
UGGGAAAGGGUGUGAAAGAUGUUAGUUGUUCUGUUGCUUGUUCUCGGUUGUUGUCUUUGG
UUGGAUUAUGUCAAAGUGAUCCCUGUUUUGUUUUCUCUAUGUCAAAGACUGGUUCUUUUC
CUCAUGUUCUAUUAUUCAUUAUACAUCAACAUGCCUCCUCUUUCCGUUAUUAUCUUUUAA
GUCGAGUUGACUAGCGUGUGACAAAGCAGACCUAUUCAGAUGGAAAAAUUGGAUAGUUUG
AUGCUAAAUAAAUCUGUACUAACCCGUUAGAGUUUGGUUUAACAGCAUACCGAGGGUAAU
UUUCCUCUGCCUUAUCCUACUGUUGCUCAUUAUUCAAAUCUAUCACGAAAUACCUCUGUU
UUUUAUGUGCGG
2. MFE structure-
..........(((((((((...(((((((.((((((...((((...))))..))))))..)))))))...((((((((((((((((((..((((((((((((((.......(((((((.((((((((((.(((((((((((.((((((...(((......)))...)))))).)).)))))))))..(((((.....(((
((.((((.((........)))))).))))))))))..((((((((..((((((..((((...(((((((((..(((((.......)))))..))))))...)))...)))).......))))))))))))))(((((....((((.....))))..(((((..((((((....(((((((((((...((((((((((...
....(((((.....((((........((..((((((.....))))))...))........))))......)))))........))))))))))(((((((((((((.(((.(((....((..(((.(((((......))))).)))..))...))).)))..)))......)))))(((((((..((....(((((((((
(((.((((........))))..)).))))))..))))...))..)))))))..........))))).........))))))....)))))....)).))))..)))))...)))))...................(((....)))..))))))))))))))))).(((((((..((.....(((..((((...(((....
...)))...)))).)))..))..)))))))...(((((((......((((((.(((.....)))...(((((...(((((((......)))))))))))).((.((((.(((((((.(((((......))))).......))))))))))).)).((((.((.........)).)))))))))).....)))))))...(
((.((((((...............)))))).)))..))))))))).(((((((..((((...))))..)))))))..........((((((((((((((((((((((((...(((((((((((((..((((((....((.(((...))).))....))))))..(((((....)))))..))))))))))))).......
))))..)))))))).....((((((.(((..........((((((......)))).)).........)))))))))....)))))).))))))...............((((((..((((((((((((((...................((((((((((....(((((..(((......)))..)))))....))))).)
)))).((((.((.......)).))))....)).)))).....))))))))..))))))((((((((((.((.((..(((((((.((((...))))))).))))..)).))........))))))))))...((((((.........))))))..)).))).))))))))))))..(((....(((.....)))...))).
..((((((((((((........(((((((......)))))))((((((((((....(((.(((((.((((((..........((((((....(((.....)))....))))))..........))..............))))...)))))))).....))))))))))..(((((((.((......)))))))))....
..(((((((...((((.((......)))))))))))))...)))))).))))))((((.........))))...........))))))...........)))..)))))).....((((.........))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-468.64] kcal/mol.
2. The frequency of mfe structure in ensemble 9.26629e-27.
3. The ensemble diversity 590.65.
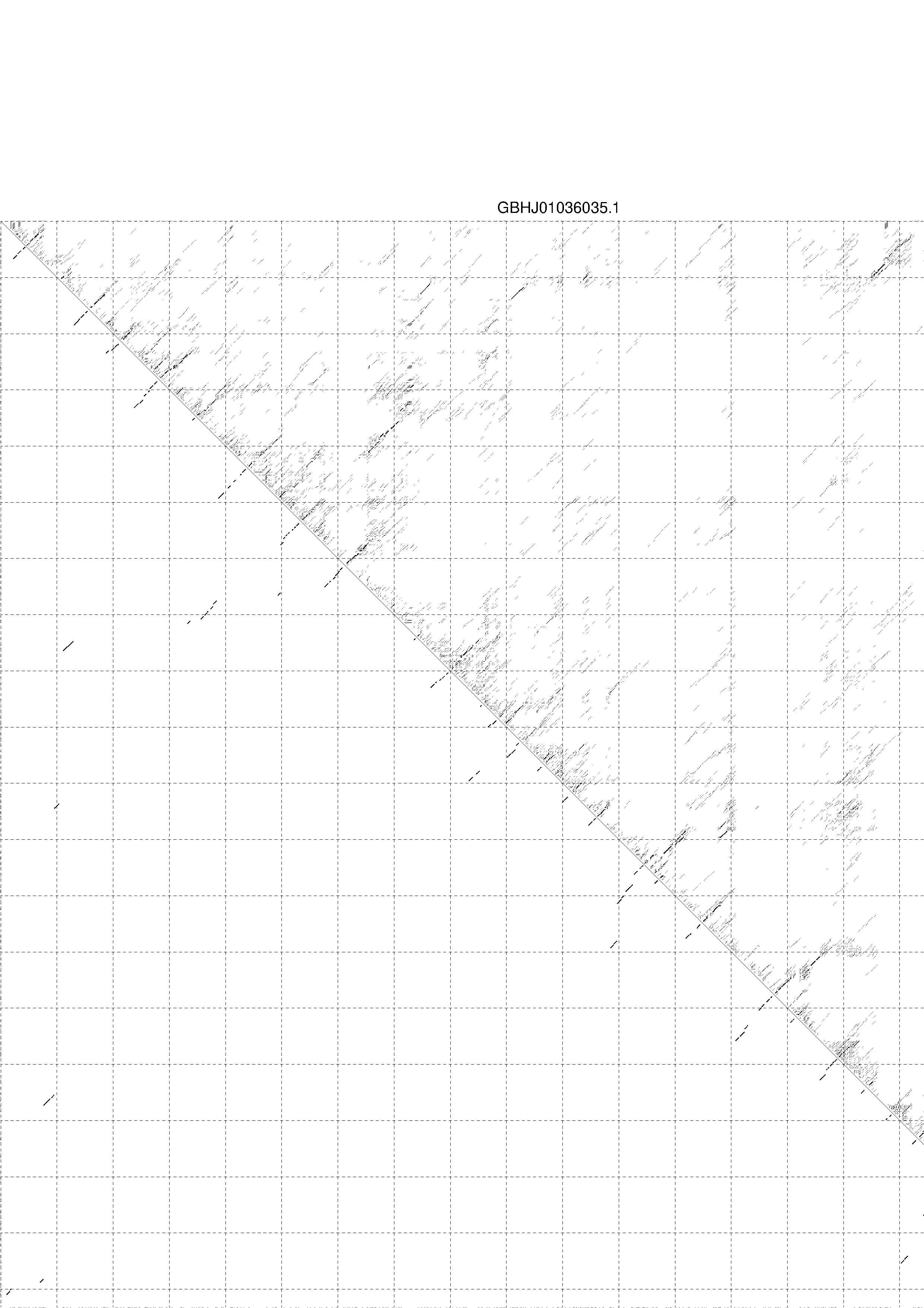
4. You may look at the dot plot containing the base pair probabilities [below]
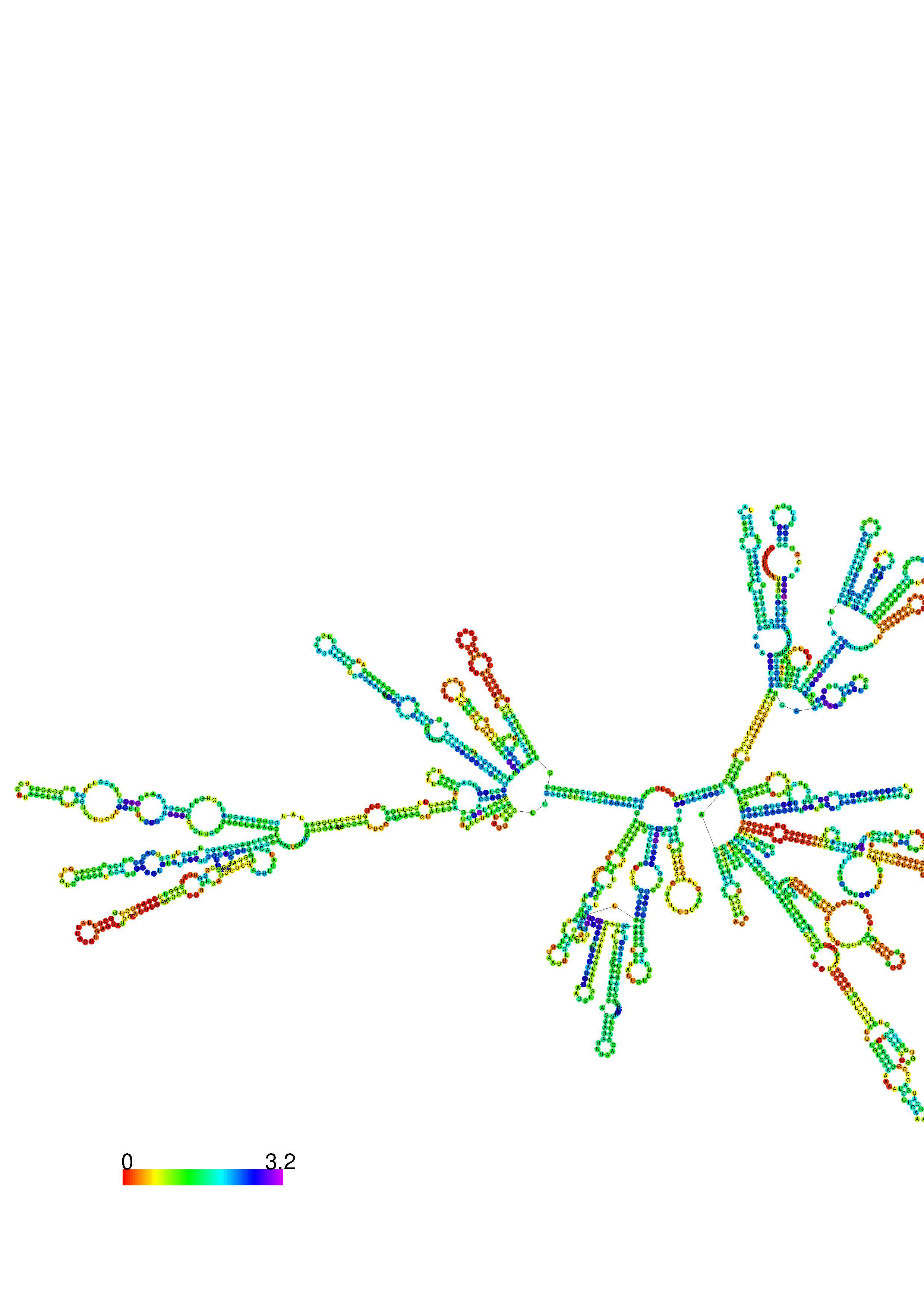
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
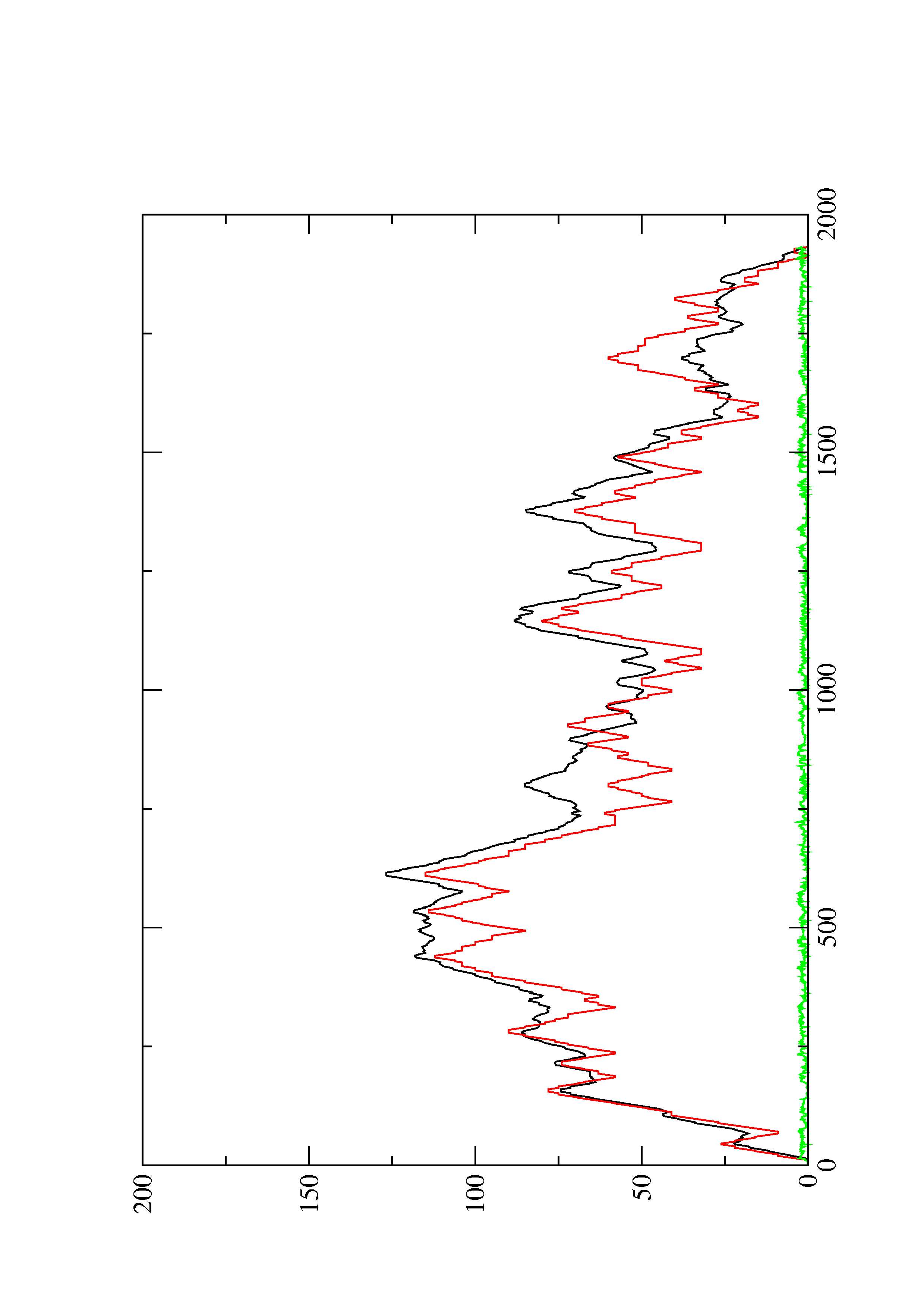
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.