lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01036980.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-549.65) is given below.
1. Sequence-
GUAAAGGAUUUCAAAUUUCAGUAUAAAACUCUGACUUUAUAGCCAAUAGGAGGUAUCUGU
UAAGAAUUUAGGUGUGCCACCGAGCCAAUCCAAACAGAGAUAUGUCGCGAGGUCAUACCA
UCUUCCCAAUGCUUUAUGUUAUACCCAACAAAAUUUCUUUUCUUCAUCCUAUGUGGGACA
AAGUCCCUUCACUUUCCAUCCAUUAGCUUAACCCAAAGGUCAUAGGAUCAAACCCCAAUC
AUACUAAAAACCCAACACGAACAAAAUAAAAUCCCUGGGGCUGCUUGAAUCAUUAACCAC
AGUGUUUAUCAACAAGAAGAAAAUGGGUCUUCAGAUACUUCUUUUCAUUUAUAUGAUUCA
GGUUAUUAUGGAUCAUUAGCUGAUUUGAUCAUUACAAAGGGCGACAGUCAGUGAGUCAUA
AACAAUGACCACAAGAACAACUAACACAACCAAGGAAUCCACAUUAAACUAAGGAAGAAA
GUUCCAAGCCUCACUUACAUACUCCGGCAGUCUCUUCAAUUAAAGAAAGUGAAAGUUCAU
AGAUAUGUAUGGUUAGGCAAACAUGUAAAUUCAGCCUAUCCUUUUAAUAUUUUGAUUGAG
GAAGAAGGCUAUAGAAAGAAAGACUCUAGUCAAGUCAGUGGUAUUAAAAGACAACACCUU
CCACUCAUCAGUAGUUUCAUAUUCAAUAGAGGGUCCCAGAAACCAUAAAGCAUUACAAAA
UCCCCUAACCAUUAUUGUGAUCUGGCUAAAAGAGAUACUUGUUAGUCAUAAAAGAGGCCA
CUACCUGAUUUUUCUGCAUGUAGAUAAAUUCAUCCUCCAAAAAACAUUUAAAAGCAAAAA
GUUAUUACAAUAACUUUCUUCUAGUGGUGCUGUGAAUUUCAAUGACAACAUACCUGCUUU
UCCUACAACUCUUUGUUAAUUUCUUUAAAUUUUGCCAAUUCAACUUCUAACUCUAAUGUA
UAGGCCGGUCAGAUCAAAAUACCACCAUGGAGUUAUUCAUAAAAAAUUCAAGUCUAUAAA
CACAUUAAUAUCAUUGUCUUCCCCUUCUCAGUCUCAAAAUUACACCCAGUUAUAACUUCA
AGAUCCUACAGUGAAAAUCAUAUACACUUCCCACAAGUCAACCUGCAAAGUCAACAGUUG
CUUCAUAAGUUAUAGAUAAUGAAUCCAUGUGUCAAAGCUCUGUAUGUAUUUAUGUUUUUC
GGAUAUUGCUAUGAUUAUCAACUUUACCUAGCAUUUCGCACAAAAUUUUUUGAAUGCAGA
UUUACUGAUGUUUGGAUAAUUAUAAAUACAUUCCAAGCUCUCUAGUAACUAGCAAAGAUG
CUCAUCCAGAGAUGUACAGAUGUUUAUGAAAAUGCAGUUAGUAGAUUCAGUUAUGUUUUU
CUCAAUUGCAUGACUAGUAGAGUUGUACAACAGCAUAAAAAAAAGAAACUUUGAAGUAAU
UGAAAGGUUCAAUAAUAACAACAUUAUUAGGAGAAACUAGACUUAGUAUGAUACUAUAUG
UCCUUAGCAUCUAAGUAGAUCAACCAGAACUCCUGCUUCUUACCAAAGUCCUUAGCCACA
AUUCAAAUAGUAAAUUGAAUGGGCACAAAUCUUCGAAAAUAUGAGUCUUAUUCCUCAAUA
UAAAUUCCAUAACAAAAUAUUGAGGAUUAGAAAUGCCAUAGAUAGAGCAAAACUCUCAUA
GAAAAUUUCAUUGCCAAAUUGAAGAUGAUUGUUUCACUCACAAUAAAGAAAGAUGCAACU
CUUAAGAUCUUACUUCACUAUAGUUUAACAAACUGAAAAACAGUAGCUAAAAAAUAGAGC
UAUUUGUACUGAUCAUGGUUUUGACAUCAACUCACUUCAGGGCAUUUUUGAAUGAGUGGG
AGACUCAAUUUUAUCUACACACUAUUAUUCAAGAUCAACAGUUUUCUUUAAAAAAAAAUU
CAAGAUCAAUAGUUGAUUCAAGAAUCAACUUCAGUCUCAAGUUUAUUCAUCCAGUGUCAG
CUAUAAGUGAAAUAUACUCCACAAUUAAUACCAACAACAUAUAUAUUAACACAACUGAGA
AUCUCGAUUGAUUAGUCUUCUAAAAGCUCAUUUUGAAUCCAGAACAGAAAAAAUUAAUCA
GUGAAAAUUAUAACACUAGCACAAUAUUUUAAUUUAAAAAAUAAAUUAAAACCAAAACAU
AGUAUCAUCCAAAAGGUUGGCAUCACACCAAAGAAGAUGUGAAAAAAUAUUCACACCUUC
CGCAAGCUCAUCUCAAUCAGGAAAAAUAAAAAGAACUUCAAGAUAUACCCCAGAAAAAUA
AGAAUAUGUGACAAAGCGAGCUCAUCUAAAUCCAAAAUGAGCUAAAAGUUUCCAAAAAUA
AAAAAUCCCAUAAAGAACAUAAAAUUAAUUCAAUAAAUGGCUAACAAGUAUCUCUUUCUA
UAAGUCGAGAUUAGUGAACGGUGAGGCGAUUAGGGUGGCGGCUGAUGGGUUUUACCACCA
CUGAUGGCUGCUGGUCGGUGGAGGGAGUAAACGAAAAGGGGGAGAGGAAGAUAGUGGGCC
GACCGGUAUUGGUGGUUAUCAGUUGGAGUUGGGGGAAAUGGAGAGAGAGUUAGAGUGAGG
GAUGUUAUUAUUUUAGAUAGAGGAGAGAGCACAUGGGAUAGGGUGGCGGCUGAGGUUUUU
ACGAAGUGAGGGGGUAAGGUUUCA
2. MFE structure-
.....((((((.....((.(((((.(((((((((.(((((((((...((((((((((((..((((.....((((((..(((..(((((((........))).))..))..)))))))))........................((((..........((((((((((((((((.(((((...))))).............
..........(((....)))))))))))....(((((......................................))))).((.((((..((((......))))....)))))).)))))))))))))))))))))))))))).(((.(((.((((((((........))))))))))).)))((((.......)))).(
((....))).(((.(((((.....)))))))).....................((...))............((((.....))))..(((...............)))..(((((((((((((((....((((((.(((((......))))).(((((...............)))))..))))))...)))))))))))
).))).)))))))))......((((.......))))(((((...................)))))..)))).))))).))))).))....))))))..((((((...............(((((((...((((.((((((..((((((((((.....))).))))))).......((((.((((((......(((.((((
(.......(((.(((((....................((((((((....)))))))).(((((((...)))).)))....(((((.((((..((((((((......((((((((.((.(((((((.........(((((((((((..((((((((((......(((((((................(((((....)))))
...........((((((...........((((.((.((((((((((.(((((((..........((((.((((....((((((((((..(((((..(((.((((.((((.....(((.((((.((.........)))))))))...))))))))))).(((....)))(((((((((((.((...((((...((((((((
((......((((((.......(((((.(((((((((((((((((....))))..))).((((((((((((((.(((((.(((((((((...((...(((((((.......((((....)))).....))))).))...)))))))))))..((((.(((.((((((((((((((((...........))))))))....)
)))).)))))).))))........((..((((.((....)).))))..)).(((((......)))))(((......)))....(((((...)))))..)))))..))))))..))))))))...(((.....)))...................(((.((.(((((........)))))))))).......(((......
..)))...((.((((((((((...............)))))))))).)))))))))..))).)))))........))))))......(((((..((...))..)))))(((((..(((.......(((.(((((............))))).)))......)))..))))).))))))))))((((((.........)))
)))..))))....)).)))))))))))....)))))..))))...)))))))))).)))))))))...........................((((...((((((.........))))))...))))...((((((((....))))))))..........((((((((.((((.(((((((.....(((..........)
))...........................(((.......((((((((((..((((....)))).(((((((((((.((..(((.................((((..........((((........((((((((((.....))))))))))........)))).(((.((....)).)))..))))......((((.(((
(((......)))))).))))....(((((......(((..(((.............)))..))).......................((......)))))))..)))..)).)))))))))))........................((((.........................))))....................
..))))))))))........))).(((.((((((((((.((((....))))...))))).))))).)))))))).)).))))..)))))))))).)))))))))).)).)))))))))).)))))))))))).)))))..)))).)))))))))))))).)).))))))))))))).)))..)))).))))).......)
)))))))...))))))))..)))))).)))).......)))))))))))))))))..)))))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-595.93] kcal/mol.
2. The frequency of mfe structure in ensemble 2.45027e-33.
3. The ensemble diversity 680.39.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
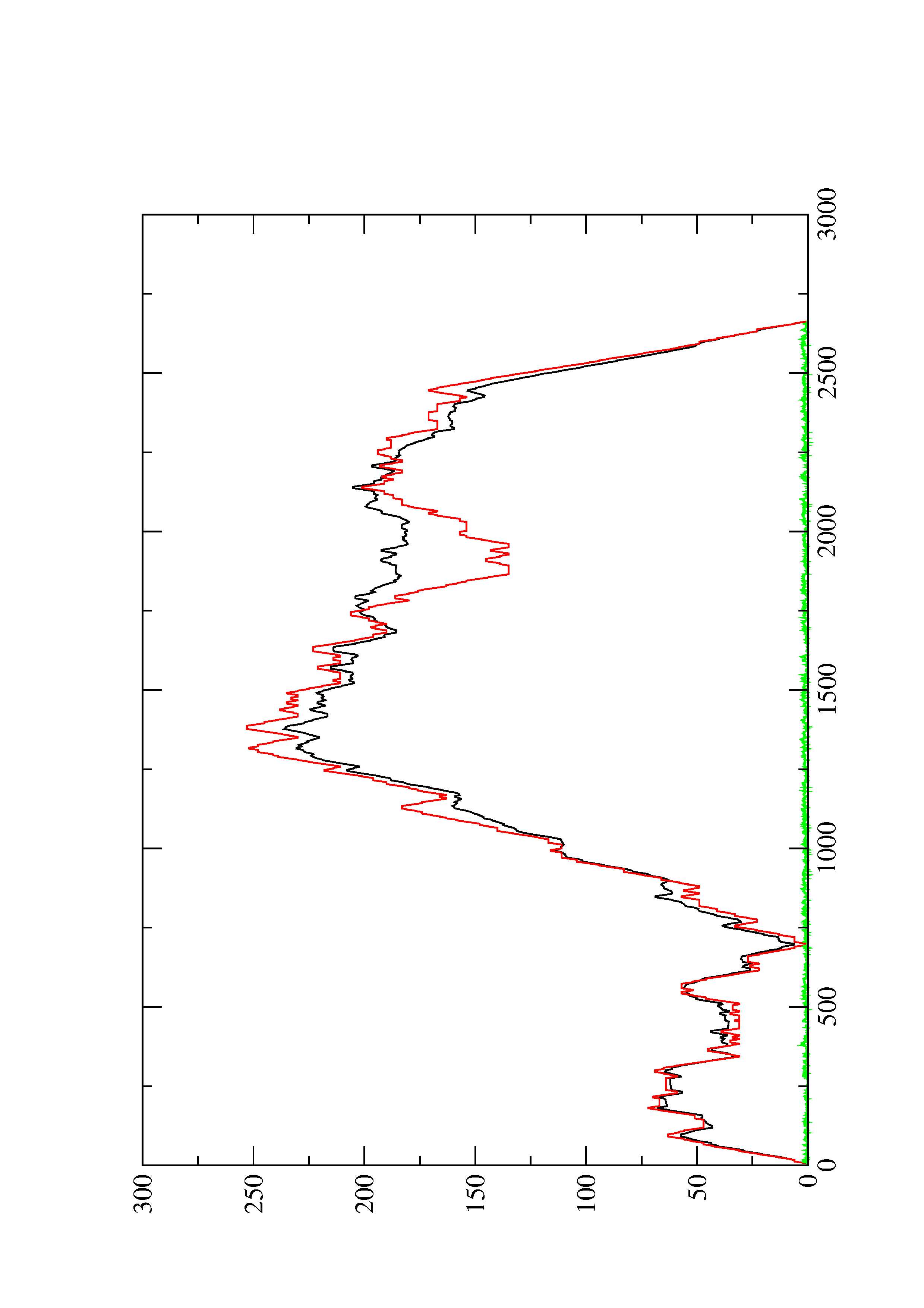
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.