lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01038133.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-376.00) is given below.
1. Sequence-
AACUUCAAUUACAGAUCAAAACCUUCACUCCAAAGCAGAAAACAUAAUACACAUCAGUGA
CAUCACUCAUAAUUACUCUUUCGAGGCAAAAAAGAAACCACCCGUAACAAGGAAGGACAA
AGUAAAACCAAAAAGCCCAUCCCGAACCAAAGAGAGAGAAAGCUGAAAGACAAAACAAAG
UCAGACAAUCAUAAAAGACCGCCAAAAACGAAAAAAAAAAAAAAAAGGAAUAUCCAAUUG
AAAUAACUCACACAAAUAAUUCUUUGCACAGAAACCACCUAUCCAAAAAAAAACCAAAUA
AGAAACACACAAAACGCAUCAAACGGGACAUCUGCACCUAAACUCAACAAAGAUACAUCU
GCCCUUCAACACUGUAAAUGGAUCCGAAAAAUAUAAUCGUAAUUCAAAAAACAAACAAAC
AAGUGGAGUCUCGAUCCAAUGUGCUCUUAGUAUUUACACCAAAGGGGUAAAAAAAGAGAG
GAACAAGUGGGUCUAUUCAAGAAUUCGAAUGUGAGGAAAUGAAGGAACAAAACAGCCAAU
UAGGUGAAGGACGAAGCAUAGUAGUCAGCAGACUCAGCACAACAGUCAGCAUUUUACGAA
CAAUGACUUGAUAAUUUCUCUUAAGCAUUUAUACUGAAUAUUAGGGAAAAACAAAAGGAA
GAAGUCUAAUAUUUAAUAAACACAAUUCCACUGUCAUUUUGUGCGAAGAUGAUACAUUAA
CAACAUGUUAUGUGUUGUGACAAAUUUAAAGAUGAUAAUAACUUUCAAGGACCCAAACUU
UGUAUUAUCCGAACAAAAAAUAACUGGCAUAUACAUAACACAUAUAUAAAUCACAAUGAA
UUGAUAGUUUUCAAAACAUCAAAAUAACUGGAUUUGAACUCAAUGAAUCACAAUGGAUUU
GAAAAAAAUUGGGUAAUGACUACCUGGACCAAAUUUAAUUAAAACUAGCAUGGCUUCAUC
ACUCUCAUAUGCUUUAAAGGCACAUUAAAAGAAAGUGGGGGAAAGACAAUGAUAGGCCUG
ACCAAAGAAGCUAAGAAAGAAGUAGGAGAAGACCAUUGGAUAUUGAGAAUACCCGGAGAA
AUUCUUUUUCUACUUUCCAAGCUCUAUUUCCAUUGUGCCAUAUCAGGGAACAAGUUAUCU
CAGCAACUACAUACCUAAUUUACCAACAAAUAAAUCAUGGAUAAAAUACUGCAUUCAAGU
UGACAGACAGCAGACUACAAGUUGACAGACUACAUUACUAAUACAGUAAUACUACAUUCA
AAGGGAAAUAGAGCAAUCAUUUUUAUAACUUUGGGCAACUACCAAACAUGGGAAAAAAGA
UUCUAAACAUGAAUAAAAUUCAAAUACCAUCCAUUAGGAUCAAAACAAUGAGAAUCAACA
AUAUUCACAACCAAGCAUGAAAGAAAAUCAACAAGCCAUAGUGUCUUACAGUUAAAACAU
UUAAAAAUCAAUUUAUGGCUAUUUUAAGAGUAACAUAACUGUGAGAAUUAAUUUUCUAAG
GAAUUUCAGCUAGCAAAAAAGAACACCAGCUAUAGUAGAGGGCAAUAAAACCAGCUAUGA
AAAGGAGUACACUGGCAGCCAUUUCACAGAUCUAGGAGAAGAAGACUACAAAAAAUAGGA
AGCCUCCGUAACUGCUUCAACUAAAAAAAUACAAAUGCUAAAAGACUACUAAAAACAUCA
CUCAAAACAGGUUCACUUUACAAAUGCAUGAGUUCACACUUCACACAGGUAUGGGAAAGC
ACAAAGCACACAGGUAUGGAAAAGCACAGUUAAUGAGCCCACGUGGAAAUGAAUAUUAAU
CUUAUCAAUGUCGGCAAGUCACAAGCAUGUAAGUAAAUUUUCCUUUAAACAAAGAAUAAC
AUCAACAAACCUCGAAAAGGAAAGCAAAAAGAAGAAAACAGAGAGGGCAAAACAAAGAAG
AUUUGAGGGAAAAAUUGAAACAAAAGCAAACUAGAAAUUGAAAGAUGAGUACUUCACUGA
UUUCAGCGUGGGAGUAGACAAAAAGAAGACGAGGGUAGGACACCAGAGUCUAGGAUGAUG
ACGAUGGUCGAGACUGCAAGAGCAGGCUGAGGCUGCUGGUCUACGACGAACGAGACGGAU
GAGAAUCUGAGAUUGGAAUUGGAAUUUGGAAUUUGGAACGAGACGGUCGAGGCUGCUGCC
CUAAAAGUUUUACCUAAACUAAA
2. MFE structure-
...(((((((...((((....((((((((..........................((((....))))........(((((((..((..........((.....)).....(((.((....................)).)))....))...)))))))...((((...(((........)))..................
..........................(((...)))...........((((((.....(((((((.....(((..((((...(((..............................(((((....((((((.(((((................((..(((.(((...........))).)))..))(((.........))).
......................))))))))))).....)))))(((((....(((((.((....)))))))..))))).)))....)))).)))....)))))))....)))))).................))))......))))))))..........(((((((((((.((((((((....((((.((((((.....
.((((.(((((.........))))))))).....(((((((((((...................)))))))))))..........(((((((((..((((((((.......((((..........(((((((((((((.....((((......(((((((.....(((((.......))))))))))))........)))
).....))).)))))))))).......(((((........(((((.((((...)))))))))........)))))(((((((...........((.(((((.(((.....(((((.....)))))(((((...............(((((((((((.(((((((((((...((((.((......)).))))...))))))
)........)))))))))((.((((((....((((((.((((((((((((((...((((.((((...)))).))))......))))))))))))))....(((((((((((.((((.((.......)).))))((..(((...(((((..........(((((((............))).))))...((((..((....
.....))..))))......)))))..)))..))((((((.....)))))).............))))))))))).....))))))...)))))).))....(((....))).......((((((.....(((((...)))))......((((....))))..........)))))).................(((.(((
((((((((..........((((.(((((((((((..((.(((((((.(((.....)))...))))))).))....)))))))))))))))))))))).......)))))))(((..............)))...((((..((........))..))))...(((.(((((...(((..((((((...((.(((.......
..))))).....)))).))..)))...)).))).))).................))))))..((....))...................)))))..))).))))).))))))))).............))))......))))))))...))))...))))).......(((..((..(((((((.((((((((((((.((
(((.(((((.((.(((.((.....)).))).......((((((((((((....((......))......((((.............................))))...............))))))))))))....................)).)))))))))).)))).))))....)))).)))))))..))))).
......((((...(((.....)))...)))))))))).))))....)).....((((....)))))))))).))))))).))))...........(((((....))))).))))..))))))).(((((..(((((...(((((......(((....)))......)))))..))))))))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-413.80] kcal/mol.
2. The frequency of mfe structure in ensemble 2.32527e-27.
3. The ensemble diversity 400.60.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
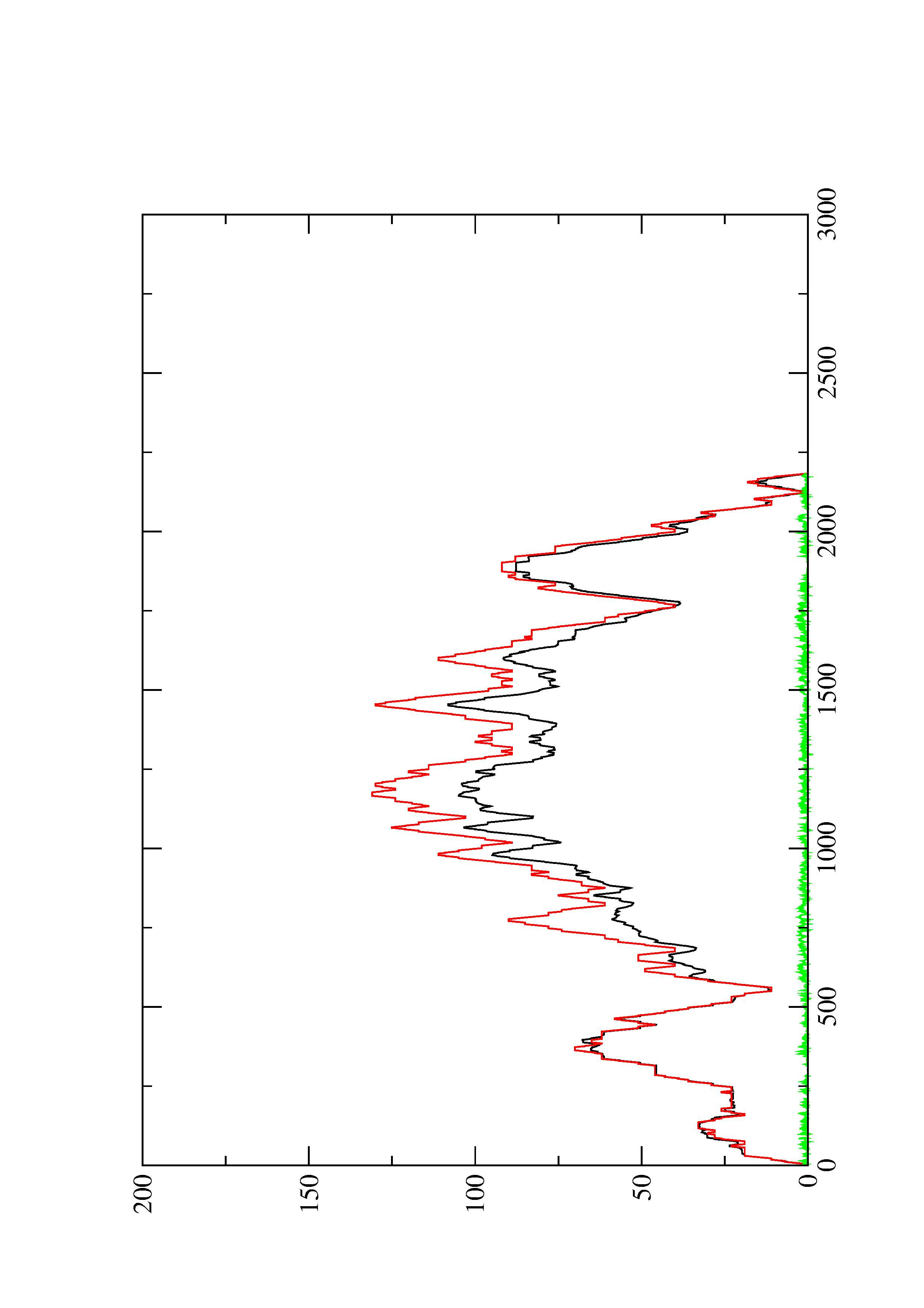
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.