lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01038396.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-583.30) is given below.
1. Sequence-
AAAAAAAAAAAAAAAAAAAAAAAAAGAAAAAGGAAGUUUCAUUGGACCAUCCGGCCCAAU
ACUUGCGAGACCGAUGCGGGGGGAUUUCGGUCCAUUGAACACAUUCGGGGAUCAUCAAGG
ACAUGCAAAAAAAAAAGUGAAUACAUUAGAAAUCAGAUAUAUAAAUGAUAUAAAACAUGU
UAGAAAAAAUAUUUGAACAACACAAUUUUAACAUGAAUUAAUCCAUAAUGAUAUCUUAAA
GAAAACAUAGCCCAACUAAGAAACAACAUUAUUAGAAUUUAAAGCCCAACGAAACAUAGA
GAUGAAACAAUGGCUCACUAAAAUCUAAAUUUUCGAAACCCAAACAUAAGAAAAGGCGAC
AUAGACAUGGAAAAAGAAAUCUAAAAAGCCAUAAGGAAUAGUGAAUCCAGUAUGUCAUGC
UCAUAUCUUUGGAAAGAAAACAAGUUUGAAACCAAUAUUAUCUAACCUGAAACUGUUUCA
UAUAUAUAUCUAAGAGAAUAGAGCCAGAACCUUUGGCACAUAGGUUAAAAAAAUAAGGAG
AAAACAUGAUGCUUUUAGGAAAAAGUCUAAGCCUGAGAUGACAAUUUGAGUACAUGGCCC
UUAAAUAAUACUAAAGCCCAUACAUGCUAGCCACACAAGCAAACAAUAUGACAUUAUUAA
GGACUUCACAGAAAGUAAUCAGGUGCACAAAACCAGGUAAAGCUAUUAUGAACAGUAAAU
CAUAUCACACAAGAGAGUAAUCAUAAACAUGCUUAGAAUAAAUCACAUAUUAGAGAUAAA
CCACCUACGAAGGGCAAAAACCAAUUAGAGCAUUCAAACUUAAAGACAUGAAACUACUAG
CUUCAAAGAGAAGAUUUACGUAGAUAGAUAUCAUUUCACAUAGACUUAAAGCUAAAUUAG
UUGACAACCAACUAAAAUAACACACACACUCAAACUAAGAUAGUAAACAGUUGCAGAGCA
GGAUCAACACCUUGAAAGGGAAUUACACGUUCAAUCAACAACAAAUAUGUAUUUAUACGU
UAUAAGAAUUUAUUUGUAUUAAAAUGAAAUCUAUUAUCAACAGGUUCCUAUUAUUAUGUU
AAAAUCAUCAAAAAAACAACAUAUGCAAGUAAGCAGAGUAUGAAACAAGUGGACCAAUUU
GCACAAAAAUUAGAUAGCACAUAGGCAGCUACCAUCUAAAAUCCCCAAAGAGAGUCCUUU
ACCAUAUCAACAGACUUGAGACUAAGUAAAAUAUAAAUAGAAUGAGUAGACAGGAAACAU
CUUAAGUUUGAAAACAGAAAGCUCUUAAUAGUAAAUCAGUUAGGCACUUAGCUUAAUGAG
CAAGAAAACACUUCCAAUCUAUUAAGAUCAGUCAUCUGGAAAAGAAAUAACAAAACCUUU
GAGGCAUUAAAUUCCCCACGUCCCUCAAUCCUUAAGACAAAUGAAUUAGACCCCAUACUG
AGCAACAUACCUCACCUUAGGCAGUCUCACAAAGUGUAAGACCUUAGACAAGUGGAAAAC
CAUUGAAUCUUACCAUAGGCGAUUAUGAUAUUAUGAACAGUAAACCAUAUCACACAAGAG
AGUAAUCAUAGACAUUCCUAGAAUAAAUCACUCAUUACAGAUAAACCACUUAAGAAGGGA
UAAAACAUUUAGAGAAUUCAAACUGAAAGACAUGAAAGUACCAGCUUCAAAGAGAAGAUU
UAGGUAGAUAGAUAUCAUUUCACAUAGAAUUAAAAAUACAUUAGUUGACAACCAACUCAA
AAUAACCACACAGGCUCUAACUAAGACAUUAAAUAGUUGUAGAGAAGGAGAAAUACCUUG
AAAGGGAAUAACAUGUUCAAUCAACAUAAAAAUAUGGGUUACUUAAGGCCAUAUAAUAUC
AACUAAUAUGUAAGCAUACAGAGGACUGGGAAACUAUCAUAUUUUGUACACAGUUAACCA
ACAUUUGCAUGCUUCAACAUAUCUUAACAUUAUUUAGAGAUUAGUGCACGUGUAAACUAA
UCAACCCACAUGUGAACAUAUUAAUGAAAGAAGGAAACAAGACUUUAGAGAUGACACAAG
GAGACACCAGUUUAAAAAGGGUUGGUAUUGACAAUGAUGUAGUUAUUUCCACAGGGUUGU
UUAACUCAUAUUAGAACUUAAUCAAGAACACCUAGGUGGUAGACUAAUUUAUUUUGUGAG
UAGGCAGUAGGGGUAUCACAGGGAAAUGUAUACAUUGGUCAGUAGGGGAAUAUAAACAUA
CUAAAAACAGCUAUUAGGCAAUACUAAAAAGAAACAUGCACUCACGCUUUGUUAAAAUGG
AUGGUCAUAUAGACAAGUUGAACUAGGACACGUCAAAUAUUCUAGAUAUCUCAGAGUUUC
AAACACAAAUGGUAUAAAUCAUGAUUAACAAAUUAAUUCUAACCCAUAUGCAGGAUUGCA
UACCACAUUAUGCUCAUAAGGGACUUAUAAAAACAUCUUGGUCAAGGUUCAUGCAAAUAU
UAAAGGAAAGGGUCAAAUAUUUAAUCCCAGGAAGCAUAUACAAGACAAGAAUAGAGGCAG
GAAUAAGGAACAAAACACAUGCAGAUAUAACAUGCAUGACAUCAUAACAAACUAAUUCUA
ACUACCAGUCCCAGAUUAUAUAGAAUCAUACAAGUGGAGUUAACCUGUAGGCACACAGUA
UGCUGCCAAGAAUUCUCAAUCUUAAGUUUAAAGCAUUUAUUUGUAUUAAAAUGAAAGGUA
UUUUCAGCAUGUUCCAAUCAUUGUGUUAUCAUCAUCAGAAAAACUACAAAUGCAGGUAAG
CAGAGCACGAAACAGGUAAACCUACUUGCACAAAAAUUUAGAUAACACAUAGGCAGCUAC
ACCCUAAAAGCCCCAAAUAGAGUCCGUUACCAUAUCAACAGACUUCAUACUAAGUAAUAU
AAAUAGGAUGAGAAGAUAGUAAACAACUUCAAUCUACAAAAGAAAGCUCUUAACAGUAAA
UCAGAUAGGCACUUAGCCUAAUGAGUAACAAAACACUUCCAGACUAUGAAGAUCAGUCAU
CUGGAAAAGAAAUUACAAAAAAUUUGAGGCAUUAAAUUCCCCAUGUCCCUCAACCAUUAA
GACAUUAAUACCCUUGGCUUCCUAGGUAAUGGUUUAGACCCCAUACUAAACCAUAUACCU
CACCUUAGGCACUCUCACAAGGUGUAAGACCUUAGAAAAGUGAAAAAUCAUUAAAUCUUG
UCAUAGGAUAUUAUGAUAUUAUGAAGAUUAAAUGAUAUCACAUAAUAUAGUAAUCAUAGA
CAUGCUUAGAAUAAAUAACACAUUAUAUAUAAACCACCUAAGAAGGGAAAAAACCAAUUA
GAGCAUUCAAACUUCAAGAAAUGAAACUACCAGCUUAAAAGAGAAGAUUUAACA
2. MFE structure-
...............................((.(((((((((.((((((((((((.((((.(((.(((...((((.((((((((((((((((.(((((....))))))))))......................((((....)))).))))))((((((....(((...(((..((((((((((...............
.....))))))))))..)))...)))....)))))).............((((((................(((((.((((.((((........(((....))).......))))...)))).))))).(((((......(((((.(((((...(((....((((..............))))....)))....(((...
.....)))....((((.(((((((.((((....))))...((((((((............((......)).((((((((((((.....((((......))))((((((...)))))).((((.(((...........((...(((((((((....((((((.(((((((....((((((((..(((((..((((((((((
((((.((((.....(((........))).((.......))............))))))))))((((......)))).(((.(((.......))).)))...)))))........(((((((...((....(((..((((.((((....((((((.......(((..........)))......(((....))).......
.......))))))..............))))...))))..)))....))...))))))))))..))))).))))))))..))))))).........(((((((.....))))))).......................(((((((....((((..((((..........(((....))).........))))...)))).
(((((((.((.((((((...)))))).)).)))))))..............)))))))......)))))).)))))))))....))...........))).))))............))))))))).)))((((...((((........(((((((((.........))))...)))))......))))....))))...
..........)))))))).......................))))))))))).......))))).)))))......))))).(((((((((....(((.((((((.....)))))))))...................))))))))).(((....)))....................))).))).......))))).))
)).)))..............(((...((..((((.((((((.(((.........(((((.((...(((((((((((.....((((....((((((....((..((........))..))......(((((((((.....)))....)))))).......((((.....(((......))).........)))).......
.....))))))...))))................(((((....(((((((.((.....((((..((((.....)))).....)))).....)).)).)))))....)))))...........(((((.....))))).......(((((..((((((((((((..........))))..))))).((..(((..((((((
...((((((((((((((.....)))))....((((((.((.....)).))))))((((((((((.((((((((((((.((((....(((..((((..((.....)).....))))..)))...)))).))))))..))))))................(((((((..........)))))))...((...((((..(((.
((((....................))))..))).)))).))((((....))))......)))))))))).(((....))).))))))))).))))))..)))..))........................)))..))))).........)))))))))))...))..)))))((((.(((......))))))).))).))
))))))))..))....)))........))).)))).))).............((((.((......))..))))....))))))))).....(((..(((.......)))..)))...((((((((.(((.((((((((..........(((((....)))))............))))))...((.((((((((((((((
((......)))))((.(((((..(((......((((((((((((((((.....................(((.............)))(((((((.....((((.........(((((((((((..(((((......((((((.........))))))....................(((((.(((......((((...
...)))).......))))))))..((((......)))).((((((...(((((.....(((((...(((((.(((......))).)))))....)))))))))))))))))))))......(((((((((....................(((......)))..........((((((....))))))............
)))))))))((((.........))))...(((.(((((.(((((.(((........))).)))))....................((((..((((...........)))).))))....((((....))))....((...(((..(((((.....))))).)))...)).......(((((((...(((...)))....)
))))))...............))))).))).....)))))...))..))))....(((((....)))))...)))))))))))((((((((((((((........))))))))).))))).))))).))).......))))))))))).)))))))..(((((....))))).))))))).))))))(((((((((((((
((.........)))))))))..))))))..........)).))).))))))))........................((((...((.......))..))))..................))))))))).)).......................
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-641.13] kcal/mol.
2. The frequency of mfe structure in ensemble 1.77642e-41.
3. The ensemble diversity 847.08.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
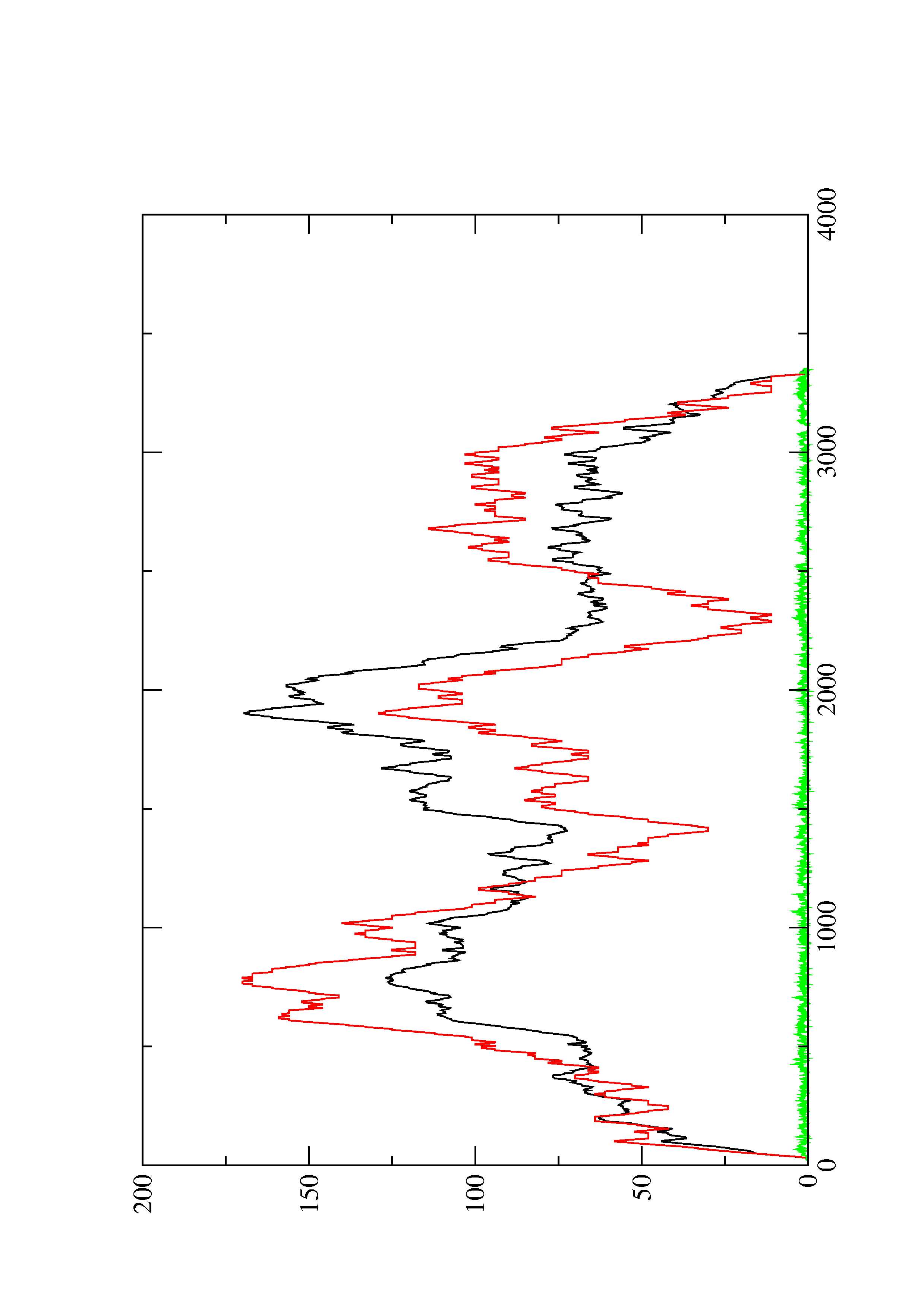
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.