lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01038515.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-637.74) is given below.
1. Sequence-
AAUAAACAUUUGUAAAGAAAUAUAAAAAAAAAGUAAAGUCAUCUCUUAGGGAACUUGACU
AAGCUAAAAUAAAAGAAUUUUAAUGUUAGCAACACGUAAUGAAAUAAAAAAUAAUAGUAA
AUCAAUAUAUUAAAUGAUUGAGCUUUGUCCCACUGCCAAGCCAUCCCGAUUUGGACUUCA
AUAUUUUCGAGCAUGUAAAGCCCAACAAUUGUCGCCAGCCCUCCGAUCUCAAUGGGUUUC
GACCCAUUUAAAUUUGUUCCUCGUUGUUGCUGCUUCGAUGGUCUUUAGUAGCGAUUGGCU
CGAUGGAUAAGUGGGCUCGUCUCAAAAGAAUGCAAAAGGGAAGUAGUCCAUGAUGAACUC
AAGUGGAUUUCAUGUAAUAGAAUAACGAUAAGAUUUAGUAAUUGCAUAUUCUAUUCAAGU
UAAAUAACAACUUCUACUAUUAUCAACAUACUUUAUCCAACAGAUUCAAUUUUCAAUAUA
UAAACUUUGGGGGGAAAUCUAAUAGGAAAAGGAAUAUAAUUCAUAUAUAAGAGGACAUAC
UGCUUACGGUAUGUUAAGGGCAUUAUUUCACUUAGAGGUGAAACAAAGCAACACAUAUGA
GAUUCAUAUAACAAACCAAAUAAGCAUGGUUCAAUGCAUAUAUAAAGGAAAUAAUUGAGG
AUCCCAGGACCAAUAAGCAUGAAAUCUCACCAGGCUAAGGUUCAAGUAAGUGUGAAUCAU
AAGGAUGACAUUUCAUAAACAAGGAAUUUAACAAGGAAAGGCCUUAACAAUACGGAUCUU
CCUUAACAGAGAUAAGACAAGUAGUUAACCCAUUCUCAAUUCCUUAGGGAUUUCAAAUGA
GGGGGUCCCUUAGGUUGCAACACAUGUUUAAGCAAUGACGAGACCAUUGACAGCCUUCUG
AGAAGAGAUAAGCUGGAAGAUGAUGAAUGUUAUGAAAUCAUCCAUCCUCUAUGCUAAAUG
UUCACAGGAACACCGUCCUUUUUUUCUCAUGAUUUCAUCACCAGCACAUAAGCAAAACAA
UAGCAAGUCAUGGCACUCAAUCUCAUAAUAGAUCAACAAUGACUCAAUUGGUGCAAGAAC
UUACUUUGAACAUAAACAAUUCAACAAGCUUAAGAAAUUAUCAUACAUAAACACUUGGAU
AAGAAUUAAUAUAGAAGCAAUUCACCUCAUAUAAGACUUUAACCUGGGAAAAACAACUAA
CAAGUUGUGCCAAGCCUCAAUUCAAUACUCUUAUUGGUAUUAGUGAAAACAAAAAGCUUA
UUGAGGGUGAUUUACUCAUUUAGCAACAACUUAAACAUUGAAUGAUACGACACAUAAUGC
AUGGAUUGACCCUUGGACCGGGACACAAAUAAAACAUCUUUGAGUGAGACAAUUCACUCU
UAUAAUCUUCCUUGAAUCACACAGAACCUGAACAUGCUAUUAGGUGCUAUUUUAAACAUC
GAACUUUUAUAUUCCCCAUCCAAAACAUGCAUAGUUACCACAAAAACAGAUUAAGAACAA
CCAGAGAAGCCUCACUGGUGGCAAAACCACAUAAAAGACCUUCAUUCAAGACAUGUUCAU
AAGUUUAGAGUGGAAAUUUGCAAUUGAACCUUAAACUAUGAUGAAUAAACAACAAAACAG
GUCUUACUUCAGUAAAAUAGGCCAACAAAUGAAUACCAGUAAUUCGUGAAAAAAAUUAAA
AUAAUGGGGAAAGUAAACCAAUGAUCAUAUACAAUAUAUUAAUGGUUCUAAUCCCAAAAC
AACAACCAGACAGCCACAAAGGUAUGAGUAAUUAACAUGCUAAAACAUGAAAGUAGCACA
UAAAUUGGAAAACCUAAUUCAAUCAAGCUUAGACAUGUCACUAUUAUCCCACUGAGGCAG
UCCCACUAGACUUCAAAGUUUGACAGGUGAUCAAACCCUUAAACAUGAAAACAAUAGUUA
AAUCAUUCUAGCCAUAAACAUCAUACUACAUACCCACUUAAGCAUCAAAUGGUUUAAAUC
UCACAUGAACAUACAUAUCUAUGCAUUUAAUAAGAGAACAGAAACCAACAUUCUAUUAGG
UCACAAAAAUAUUGCUUUAUACAUGGUAGAUAGGGUUAAGAUAGCUAAGUCUCGAACAAG
UUACUAUACUAAUUUUAGCAUGAAACUAAACCACAUUCUACAUAAACAAGUCCAUUUACA
CAACCAUGCAUCAGACCAACAACAAAUAGCAUUCAAACACUUAACCCUAAUCAAUAAGUU
CAACUUAAAUAGGCUCUAAAACAUGUUGUUACAGCCAUAAGAAGGAAAGCCCUUAAACAU
GAUGGUACUAACCUCGAAAAGAACAUUCAAACUAACAAAUUUGAUGAAUUAAAAAUUCAU
GACCAUUAAACACUCCAAAUUUUACAUAAAACAAUGAUUGAGAUACUCAAAUCCUAACAU
GGUCAUUAUAUAGUGAUAUUAACAUCAAACGAUUUAGUUUGUAGCGAAACACACAUACUU
AUUCAUUCAAAUUGCAACAACAGCAACCAGUACUUCUUUUUUCUCAAACCAAUAGAACGU
AAAUGAAUAGAAGGACAAAUCUCACCUUUUAUUGUACAGAGACAGGAAUCUAAAUGAGUU
GAGAGUUAUUUCUAUAGCUUUAAGGCCCUUUUUAAAGAAAACUCAAACCUCUCCGUUUCU
AAUCACCCAAUAGAUAUAAGGAAAUGCACCUUAAAAUGUCAUAUGGAUUAUAAAUAACUC
AUAAUUUUUACCCAUCUUUUAUAAAAUUUGUAUUCUAUGACUAUAGAUCACUCAGUCACC
CUAAAAUUCUCUUAUAGAAGCCUUAAUUUCUACUAUUUAAUCAUUUAUGACUAUCGAUGC
CUUACUCUAGCUCACUAUUCCUUACCCCUAAUGAAUAUCUAUUAUAGGGUUUAUAUAGGA
AAUAAAUUAGGGUUCAAGAGAGAGAGAGAAAUAGGGGAAAACCUAAUUCUCAGUCAGAUA
AGACCAUUAGGUGGCAUGAAUCAAAUACAAUAGUACAAUCGUGACCAAAUUAGAUGGGCC
AAAAUCAAUUUAGGCUUCCAAAUUGGAAUUUUUGAAAUUGGAGGUUUCAAUCUCACCAAC
CAGAAAAUAAGUAGUAUGUGCUAUAUAGAAUGAAAAAUGCAUUGAUUAAGGAAAAUUCAA
GGAAGAAGCAAGUUAAUACCGUCGUUGAGGACCUGAAAGGUUAAAGAAUUCAUCCAACUU
AGCCAUAUCAUACACAAAAAGAGGAAAACAGAUGGAAUUCGAUGAGGUUAACAGAAUCAA
AGGCUUUGCUCGUAGUUUUGCACUGAUUUGGUAAGGAUUUUGGGGUUUCUUUGUACUCGU
AGACGAAAUAGUUUAAGGGGAGGGAGUGAGGCGCGUACAGAUGCUCUCUUUUG
2. MFE structure-
........((((((......)))))).....(((..(((((((((...))))...)))))..)))....(((((((((((((((((.......))))..))))))..............(((((..........)))))(((((((((((((((.((((((((........))).)))............((((((...(
(.......(((..((.(((((.........(((((((....)))))))...((((((((.(((..(((((((((............))))))))).....))).))))))))))))).))..)))......))..(((((...((((((((((((((.((.....)).)))))..(((((((((.((((...........
))))..))))))))).(((((.......))))).(((((.(((((.....((((.(((...(((((...(((((((..........)))))))..)))))....)))))))((((...))))....((((((((((((((((....)))))))))....((.((.(((((((....))))))).)).)).....((((((
...))))))..........(((.((((......)))).)))...........((((((((.(((((((.......(((.((.........)).)))...((((((((((((((((((((((((((((..(((((((((..((((((((......((((.((..(((((..(((...((((.(((....)))..))))...
)))))))).)).)))).))))))))(((((((((.......)))))))))...((((((((....)))...)))))........((((..((((..(((......)))...))))...)))).......)))))))))....)))))).))))......))))))((((......))))........((((((((.....
...((......))...........))))))))((((.((((.((((............))))....))))))))....)))))).))))))..................((((....(((((...((........)))))))...))))....(((....)))...................)))))))...((((((..
..))))))......)))))))).....)))))))))))).)))))..........(((...((((.........))))...)))...........(((((..((......))..)))))))))))))).)))))(((..((((..................((((((......)))))).(((((..(((..((......
..((.((((((........)))))).))........))..)))...))))).))))..)))...))))))...(((........)))..............(((((((((((((...((((.....))))....(((((((............(((((((.(((((((.((...((((...))))..)).)))))))...
)))))))...........))))))).......................((((((.......))))))...................((((.....(((((.((((.((((...)))))))).)))))....)))).........(((((((((...(((((.((((((((((((((.((((...(((((.....(((...
..((((((.....)))))).......)))....)))))........(((...((((....(((........((((..((((((..(((......))).)))))).))))..((((.((....((((((((...(((..((.((((((((....................((((((...((((...((((((((......)
)))..))))....))))....)))))).((((..(((.((((((........((((........))))((((((((((((((.((((.((((.((...(((......))).......(((((..........................................))))).))))))..........)))))).......)
)))))))).)))..(((((...)))))....((((....((((...))))..)))).....((((.....))))......(((.((((((...((.....)).(((.((((........)))))))........(((((((.(((...................(((......))).(((((...)))))..((((..((
((((.((((...(((..........(((((......)))))..........))).................((((.......))))...........((((((((......((((..(((((((((((((((((..........)))))))))....((((((.(((.......((((((.((((((((....)))))))
)......(((....)))..))))))......))))))))).(((........))).(((((.......)))))....((((((.((((.((((((..............................))))))))))))))))....(((......))).................((((((......))))))........
.)))))))).)))).((.((((((...((..((.((((.....((((.(((((......)))))..))))....))))))..))...))))))))..))))))))(((((.((((......)))).)))))......)))))))))))))))))))))))).........)))))).))))))))).)))..))))((((
.(((....))).))))((((...)))).((((((......(((((....)))))......))))))....)))))))))).))).)))))))).....)))))).....)))...)))).)))...)))).))))))(((.(((((((((((((...)))))..........((((.((...((................
..)).....)).)))).)))))))))))..................)))))))).)))))..))).))))))........)))))))))))))..(((.(((((......))))).)))..)).)))).)).....))))).)))))))))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-702.58] kcal/mol.
2. The frequency of mfe structure in ensemble 2.04256e-46.
3. The ensemble diversity 928.74.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
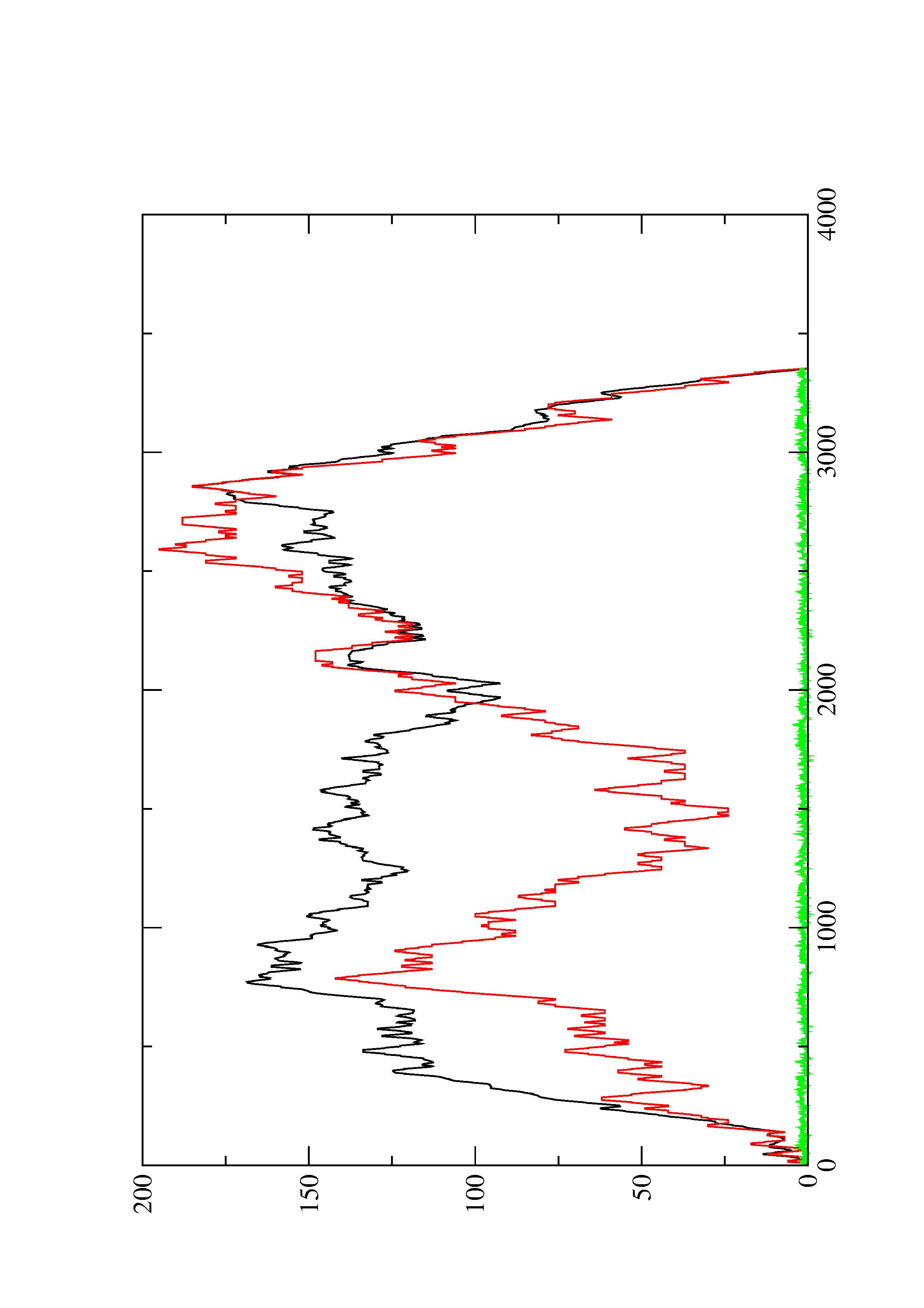
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.