lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01040945.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-439.30) is given below.
1. Sequence-
UCCACAGGUUUCAUUAUGAAAUCACUGAAAUAAAAGCAACAGUUCAUUAAACAUUUACAC
UACAACAUAUUGAAAAGUUCAACAUCAUGUUUACACAUUGAAACAUGCAUAUCACUUUAA
AAUCCUAUCCUAGAACAUAUAUAAGUUGACAAGUUUUAAAAGACUCCAUUCAAUAGCUAG
UGAUGAACGCACAAAAAGUUGUAUGCAAGACCAUUUGCAAUAACAAGAUAGGCAAGUCCA
UAGUUAAUUCUAUUAUUUCUUUGUUAGGUUGGACUUAGCAUUUCUUUAGUCAACUCAGGU
AUCAUUUAGCAAAUAAGAAUAUUCCAAUUUCAAACAAGUUUGCAUAGUCUACAAAUCGCC
CAAUACAACAUACAUUUGGAGGGUAGGACCAACGAAUAAGCUUAACUAUAUUAAAGGGAG
AUCGUGAUUGUUUAAUUGAUAAGACAACAUACAUAAGGGCAUGGACCAGUCAUACUCAAU
CAUGUAUAAUUAUCAUAUUAUAGGUUAGAGGGGUCUGGAAUGGAACAGGGAUUAACACAU
AUGCCUUAGUCAGUCAACAAUAGGCUUCAUUUAAUACAUAAGUAAACAAAAUCAGCCUUA
UACAUCAACCAUUAGAUCUUUAAUCACAUAUGGCUGUAAUGUCUCAUUAGACAUCAAAAU
AGUACUAGACAACAAUACAUCUGAUAAGAACGUAGCCAGAUUUAAUAAUACGAGGGGAAA
AUCACUUAAGGUGCAAGAAAACAAAUGACAUGCUCAACUAUCAAUCAACCCAAGGAUAAA
UGUUGCUUAGAAGGUAUACAAGGCCACAUAAGAAUCCAUCAGCACAAAGACAGGCAGGUA
GAUCAUGUAAUAUGUUAUUAGUACUUAUUCAUACUCAACUGUCAACCUAAUGAUCCUACU
AAUAGGUUCAGGGACCCAAUGGAUUAAUUAGAGCAAGAUAUGCUAGUGAAACAAACAGAA
UUAUUCAGGCUGAGAAGUAAUAGCUAGUUAAUAAACUUAUAACAUAUAACAUAGUUGACA
UAUAAGAAGUUAUUACAAUAUUAAGAUUCGUUAGUGAAAAUGGGUAGGCAUAUUAAAUAU
AAAUUUGCAACAUCAGUAUCAUAUUGAUCUAUUUAUGAAGCCUGCACAACAUUGAAAAAU
AUGUUAUAAAAGUCAAUCCUAUACAAUCACCUAUGUUGUCUAAACCUGUAACAUUAUUUA
UUUAAGUGCUCUAACUUUGUGAUCUUUAACUCAGUUGGGUGAUUUCUCAAAAUUAAUUCA
GCCUCCAUAUAAGGUCGUCUCUAACGGCGUAUAUUGUAUUUUAAUACAACAAGAGGUUCG
GUCAUUUGUUAAAGUCAUUAGUAAUAUCUGAUAACUCAUUUUUGCGAAUGAAAUCACACC
AAUAUAGUAACAUAGAGAAAUUAAUAUCUGACCACAUGGAUGCAAGAUUGCAAUCUCCCU
AGCUUCUUUCUACGAUUCUAGUCAAGCCCCGAUAAUAUGCCUCGAGAGCCUGAGCAAUAU
CAUAAAGUUUGUAUAUGAUACAAAAUACUAAUGUUUGGAUGAGUUUGCCAUACCUCUGUC
CCCUAUGUUAGCUAGCCAUGCUCUUAACUUUUCAAAAAUGUUAGGAUGCUUGUCAGAUAC
UUCAAAAGUAGCGCAUUUUUGGAGGACCAAGAUGUGGAUACGACUAUAUUUUUGAAGACU
UUAAGAAACUUAUCAAAAUACAUACUGACAAGUAGCAUCAAUAGAAAUAAUUGGACAAUG
CAGGACUAAGAGAAUAACAUAAACCAACAAAAAAGAUGUAAUACAUACCUUUCUCCCUCC
UACAAAUUGAACCAAAUUUAUCACAUAUGCAAUGAAACUGCUGCACAUGAGAAUUUCCAU
CCUUAUAGAAAGCUUGCAGAUAAAGUUGCAGCAGCCGGUUUUUAUGUAGUAAUCCCUAAU
CUCCUCUGUGGAUCUAGUUUCGCCCGCGACUAUGUGGGUCUUCGUCAUUGCAACAGAAAG
AGGAGAAGAAAGAGGAGUCAGAGAGAUUAGAGAGGAUUUGGGACUUUCAUGGUUGUGGCU
UUCCGUGAGGGCAUUGCCGAUGGCAGGGUGGUGGAAAUCAAAAAUGAGGCAGAGGAAAAA
GGGAUAGAGAGAGAGGUGGUGUUGGACGACUUGGGUUGGAG
2. MFE structure-
((((..(((((((...)))))))((((............)))).................................((((((((((((((((........))))))).....((((((....(((((((((.............((((((.(((((....)))))..............(((.......))).....(((
((.(((((......))))))))))((((...(((((((((...(((((..............)))))..))))))).))...))))..))))))((((((.((.....((((((.......................))))))................((((............)))).((((..(((....(((((((
((............((((((((.((((((((.((((((((.....((((((....((((((((..(((.(((.((((..((((.((((((......)))))).))..)).)))).)))..)))................))))))))..(((((......((((((((((....(((....))).......(((((((..
........((((..(((.....)))....))))((((..(((.(((((((.(((.......(((((.............(((....))).((((..............)))).(((........)))..)))))................(((((...((((............)))).((((.((((............
)))).))))...(((((((.........(((((...((((((....(((.((......)).)))....)).))))...)))))..(((..(((.((((....)))).)))))).....))))))).....)))))....)))))))))).))).))))........)))))))...(((((((((..(((...((((...
............))))......)))..))))))))).........(((((((......)))))))..(((.(((.......))).)))...(((((((...)))))))......))))))))))......)))))....))))))......))))))..)).))))))))((((((.(((.................(((
(..((((......)))).))))(((((((((..(((((((((((...............((((((.(((((..(((((.....))))).)))))(((((....)))))....))))))...((((((((.(((((.(((((......))))).)))....)).)))))))))))))).)))))..)))....))))))..
.........))).)))))).((((....))))))))))))))))).........))))..)))..))))(((.........))).))))))))((((..((((....(((((((...)))))))..............))))..))))....((((((((.....((...((((.((.((((((((((...........)
))))))((((((((((...(((((((((((.((((((((.((....))))))))))..........)))))))))))....((((...))))............))))))))))...............(((((.((((((((((....(((((............(((.......)))...........)))))..)))
)..(((((......))))).((.(((.((((..........)))).))).))........((((((.((((((((((((......)))))(((((((........((...(((((((((((((.((((....(((..((((((((((......)))))).(((.((.((........)))).)))..))))..)))..))
))))))))))...)))))....)).....)))))))))))))).))))))))))))))))).))))).))))......)).......)))))))).....))))))).))..))))))))))))))).........)))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-482.55] kcal/mol.
2. The frequency of mfe structure in ensemble 3.32488e-31.
3. The ensemble diversity 556.82.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
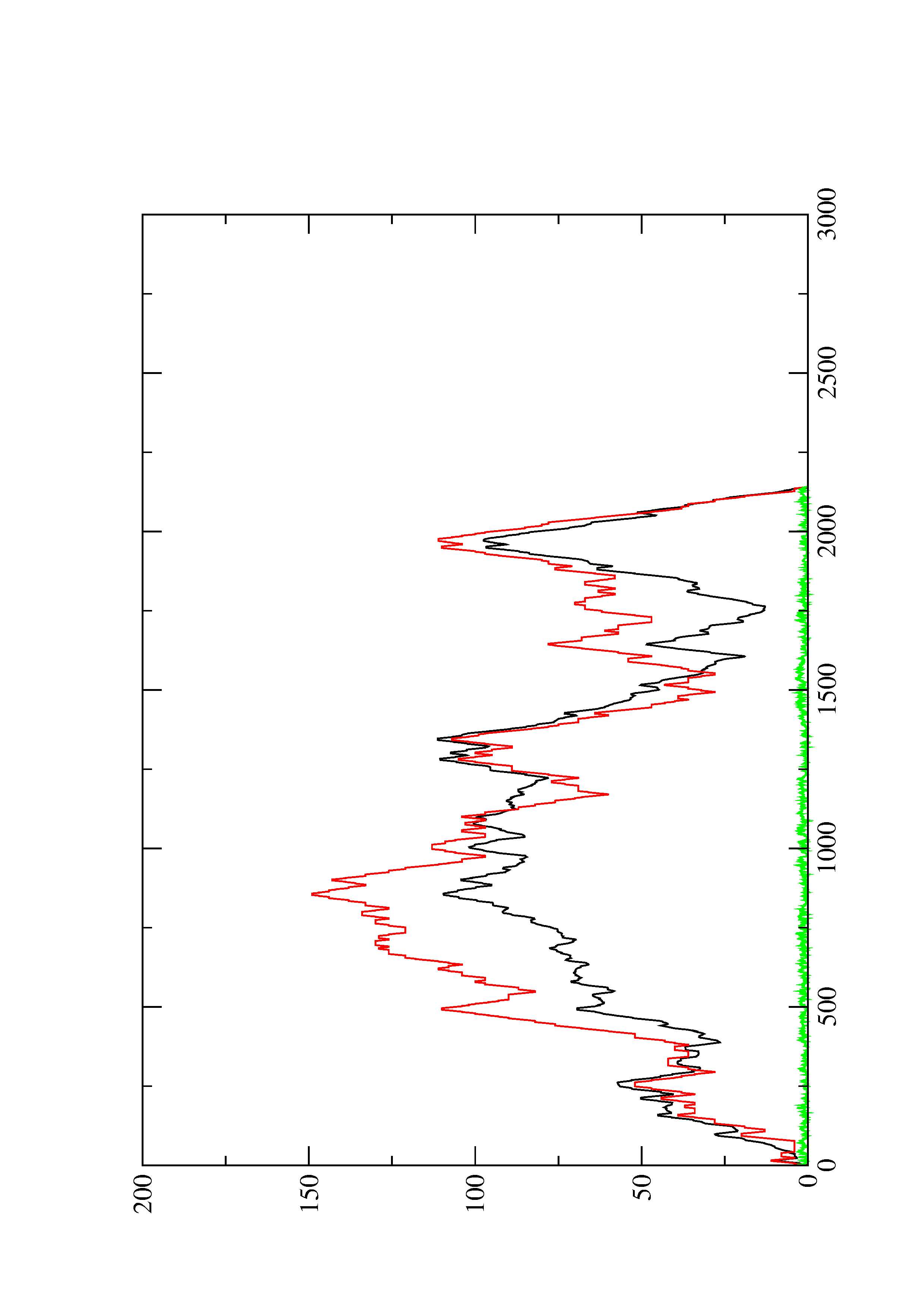
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.