lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01043041.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-636.60) is given below.
1. Sequence-
UGGUAGUAUAUUCCUUGUAUGAUUCUUCUCUUAUUUAGAAGUUCUCUAUUAAGGUAAAUA
UUAAUUACCUUGUUGUGUCGUUAUUUGUGUGGGCCACUACUAUUAGGCCCAUAUUAUUUA
GGUGGGCCCCAUGUGUUCUUUUAGCCACAUUUCUACAAUUAAAAAACCUACUGAUGAUUC
AUCCAAUUCACUAUUUAGUACUCCUUUUUAGUAUCAAGGCUUAGCAAUUAAAUAGGAAAU
ACAUGAAGCAUGAAGCGCAUAAUUAAUUCAAUUUUCAAGAAGUACACGACUCCCCUCAAG
UGAAGACUUAAAUAUCCCCAUGGAUAGAUGCAUCACAAACAUGCUUAUUACAAUUUAAAA
CUCUAGUCUAUGCAUUAGCCCUAAGCAACGAGAUGAUCAUUAUCAUACCAAUUUUCGAGU
AAAAGAAACCAAGUAUCCCUAACACAUACUCUGGAACUAUCCAUAACCACUCAUUAUACA
UCUCAAAAGGUAUUAUAAGAUAAUCAUUACAUGAAAUCAUAUGAAUCCUAUAAGAAAGUU
CAAGAUGAAUAGCAUUUACCUCCAUGGUCCUAUAUCAGGGUCAUCUCAUUUCCAUCUACA
AUUAUCAUGCUGGCAGUAGACAAGUGUUACAAAACAUAAACACAAUAAGCAGAUGCAGAU
GAUCAAAUUGAAGCAAACUUUACACAUUUGGCCUAGUUAACCAUAUACAUAUAUAUUAGA
AAAUCAUGCCAUCACAACCAAAUUUAGUCGAAUAAGGCUUCAUCAAUGAACAAAUAACAA
CUUUUUGAAAGACUACUCCUAUCCGUUAUUUCUGGAAAAGUGCAAUAAACACGAACUACA
CUUAAUCAAUUGUUAUGUACAAGAGCAAGGUUCUUAAUACAUGUCUUAUCCAUACUUAAA
UUAGACAAGCUUUAGGUAUUUAUGACGAAGUUUUAGGUAGAUAAAUUCUCAUUGACCACU
ACAGGUUGUAUAUAAGCUAGGAGAUCACACAAAUCAUGCACAAAAGGCUCAAGAUGAGGU
AGCAUCUUGAGUGAUUGAUAUGUAAUUGACAUAAAUACAAAUUCCAACUACACUAGUCUC
AUAUGACAUCUUAAGCCAACUGUAACAUUUCCAGAUAAAUUGAAGACUUAAAAUAUAAAU
CUUAUGCAUUACAAUAAAGAUGACACUUCCGGACAUGUUUUACGCAUGAAAUUGGUCUCA
AAACUCACAAACAAGUUACCUUUUCUAGACAUACAUUGCAAACUCACAUUUUUUUCAAGA
UAACUCAAUUCCUUCUAUCACCCAAAUGUUCAAAACUGAAUUAUAAACAGAACUCAAACA
UGGUUCAUUUUUAUGGUUACUAUAGGAUGAAAAUACAUUUAGUUUCUGAUCAAAUAUAUC
AACUCAAGCAACAAGGAUUGCAUUAAACAAUGUAACACUUCACUCGGAAAAUAUUUUAUG
CAAGAUCCACCCCAAAAUUCUCUUACCUUAAAAUGGAAUCCAUAGACUAGAAAAUUGGAU
CAUAGGCAAUUUUUAGCAAGGAAUUUCAGCUUACAUUAAAGCUAUGACCUUUUCAGGGAA
GUUGUCAACUUCUUUUAAAAAAAAAUAGGAAUUAGAAUUUGAGACUUAGUCUAUUACACC
UCAAAAUACUUAAUCAUACAUAGGCUGAAUACAAGUACUUAUUAAACAUGGAAACUAAAU
UAAUUAUCAGCAUCAAGUAUUAGUCAGAAUUAGGAUAUCAUGAAAAAAAUUCUUGACUAU
CAAAUAGAGAUAAGGCUAUAAGGGUGUCAUAGGUACAGUACAAGAGGUUUAGACAUCACU
ACAUAUGAUCACCAUUGAUUUUAAUCACAUUAUAGACAUAAAAACAACUAAGACUACACA
UUGGACCAAACUGAAUUCAAAAUAGUAUUACAACCAUUCAACCACUUCAAGAUUCUUUUA
AAAGAAGAAACUUAGCUUAAGGGUCAUGUCUACCACAUUUACCUCACAUUCCCAACACAU
UAAAACAAUGUAAGUUUAAGAAAACAUUUACACGUCCAUUAGUUCCCCCUUCCCAUGAAA
CACAUUUGUUUUCAUGUAAAUUAAAUAAGUUACUUUCUUGAUCACAACUUCAAGCUCUAU
AAGGAAAAUUAGACAAGUUCACAAGGGAAUAUUAAGAGAAAAGUUUUAGUUUUAGUAUGU
UGUAUUAAGCCUAACCUCUAUGUGUAUUGUACUAUGACUGAUUAAAACAAGGCAUCAAAC
UUCAUUUUUGAAUCAGUCAAGAGAUUCCAAAAGUUUCAACCAACAUACUACAAAGGAAAC
AGGGAGGAUUAGACUUUCAUAUGCAUACCAAGCACUCAAAGAUAAAAAGAACCUUAAAUA
UCCAACAUUCAACACAAAUAAAACAAGGAAAAUCACAUAGGCUAUUACCCUUCCCAUACU
UUAUUAUCUAGACAAACCAAAUAGAGCAUGAAUCCAACCAAAACCAAACUAAUUUUGUAA
AAGAAACUCUAUUAAACAACCUCUUUUGCUUUAGAUAGCAUACACACUAAACAAACAUUA
AUCCACUCAUUUGUCAUAAUUCUUAUUCCAUUUUUCUUUUCAUUUAAAAUAUAGAAAACA
CAGACAAACUAAACUUUUAAAUUCAUGUAAAGAUACAUAUGGAAGAGAUUUAUAUCUUUU
UAGGGGAAAUCAACUGAGAUGAGAUGAAUUGAGACGUAUUUGCCAAUGUUUUCAAAGUUU
UUUAAAUAAAAUUAAACCUCUCAGUUAUUUAACCCUCGUAUAAAUACAAGGAAACCCCCG
UUUAAAGAAACGUCGUUUAAGAUCACUCGGAUUUUCCGCGUCGAUAAGAAACUUAUAUCC
UUAACUAAAAACCUUAAUCGGCAAUCUAGGCUAGAACCUUAUAAUAUAUUACUCCCUAAU
CCCUAAACCCUAGCUUCCUUAAAAACCCCUAAUUUCCACUCCUCCCUCCUUAGAACAAUG
AAGAAAUCGGGUAUUUAUAGAAGGGAAUUAGGGAAAGAUUUGGAGGGGGGAAAUUGGGGA
UUUUGAAUUUUAAACGGGAUUCCUAAAAUCUCCAAUUAGAAACCAACAAGAAAGGUGUUA
UUUGCAAAAAAAUAGAGAUUAAAGAAUAGAAGGUUACCCUUAAAUGUACACCUGGACAAU
UUCAAGAUGGUUCUAAAAGCAGACAACUAAGGUUUUGACGAAGAAUGGAUGGGAGUUAAC
AAAGUUAAUGGAAUAUUUGAAACUCUCAUCAUUUGAAGUUGUGCAAGGGACUAUUUAGAU
GAAGCUUUGACGGGGAAGAGAAGGGGGUUUUAGGUCAUCUCUAGAAGGAAGCUUCAAGGA
ACGAGGGAGUGGUGCGGAAGGGCAGGAGAGUGGGGCAGGUGUGGUUUCUUUAGGUAAAAA
UGAAA
2. MFE structure-
((((((.((((((((((..((((((((((....((((((((.(((.....(((((((.......)))))))((((((...(((((((((((((((..........))))))))).((((((...((.((.((((((.(((.(((((..(((((((......................(((((....(((.(((((((((.
((((((.........))))...))..))))))))))))....)))))((.(((.((..((((((((((...((((((((((((..((((((.((((...((((.........(((((....)))))......)))).....(((((((..((((((.....(((((((((((.(((.....((((....((((.((((..
...((((....(((((((((...((((((..(((((.........))))).((((....))))((((((.............(((((((((.(((((.(((..((((...(((....)))))))))).)))))....(((((.(((((.........(((.....)))..........(((((((((((((...(((((.
...............)))))...(((((..........))))).......))))).))))))))....(((((....)))))......((((..........(((((....)))))...........))))..............((((......)))))))))..)))))........)))))))))............
....(((((((.((((((((((((((.............((((((....(((((((((((.(((((...((.......)).(((((................))))).)))))..))))..)))))))....))))))(((.....)))....((((......))))((((.....(((((((((((......((((((.
(((...((((((((((......))))))))))..))).(((((.....)))))......................(((......)))..............(((((((.(((.((.......((((.(((.....))).)))).((((((........)))).))..)).))).))))..)))))))))...))))))).
.((((........)))).......))))..)))))))))).........))))))))))))))).......................(((((....)))))........((((.((....)))))).......)))))).....(((((......))))).)))))).........................(((..(((
((((....)))))))..))).)))))))))......))))...))))...........))))....))))......))).)))))))))))))))))....))))))).........((((......(((((......)))))....))))...)))).))))))..))))))................(((.((.((((
(....(((((((((.........................)))))))))...))))).)).))).....))))))...))))))))))..)).))).))..(((((((((((................))))).)))))).....)))))))..))))).)))(((((((((.((..(((....((.((....)).)))))
)).))))).)))).(((.(((((.((........))..))))).)))........)))))).)).))...))))))...))))))...))))))...............)))..)))))))))))))))..)))...))))))..))))))))))..(((((((.......(((.(((..........(((((((.....
.....))))))).((((.....((((((((((((((((((.((....))))))))).............(((((((((((.((.....((((.(((((((..((....(((((.((((....(((((((((.(((((......)))))((((..((((((......(((.(((((((((.(((...(((((...((((((
(((((((.(((........)))...))))).)))))))).((((((.....))))))...........))))).....))).))))).)))).)))......)))))).))))..................(((......(((.......................))).........))).)))).))))).....)))
).....))))).....))..))))))).))))....(((((((((....(((((((((..((((((((.....(((((........((((......)))).........................((((((((((((((.((((.((((((...(((((.(((((((((..(((..............)))...))))))
....))).))))).....)))))))))).)))).((.((((((..........)))))).)))))))).)))).............)))))....))))))))..)))))))))..((((((...........(((.(((....))).)))....(((((((.((((...((((((((...(((((.((((...)))).)
).)))...)))).................((((((..((...((((((((......)))............((((((..(((((((..((...((((..((((.((((...(((((...((.....((....)).....)).))))).)))).))))..))))))..))))))).........))))))..(((((((((
(((((((((((....)))))))..)))))))))))))....(((.........))).((((((......)))))))))))...))....)))))).....((((((....((((.((((((((((....(((((.....((((..((....))))))....))))).((((((((((..((((............)))).
))))))))))..)))))))))).)).))...))))))))))...)))))))))))..))).))).)))))).))).....((.((((....)))).)))).)))))))))))...)))))))..))))....))))..)))))).......))))))).......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
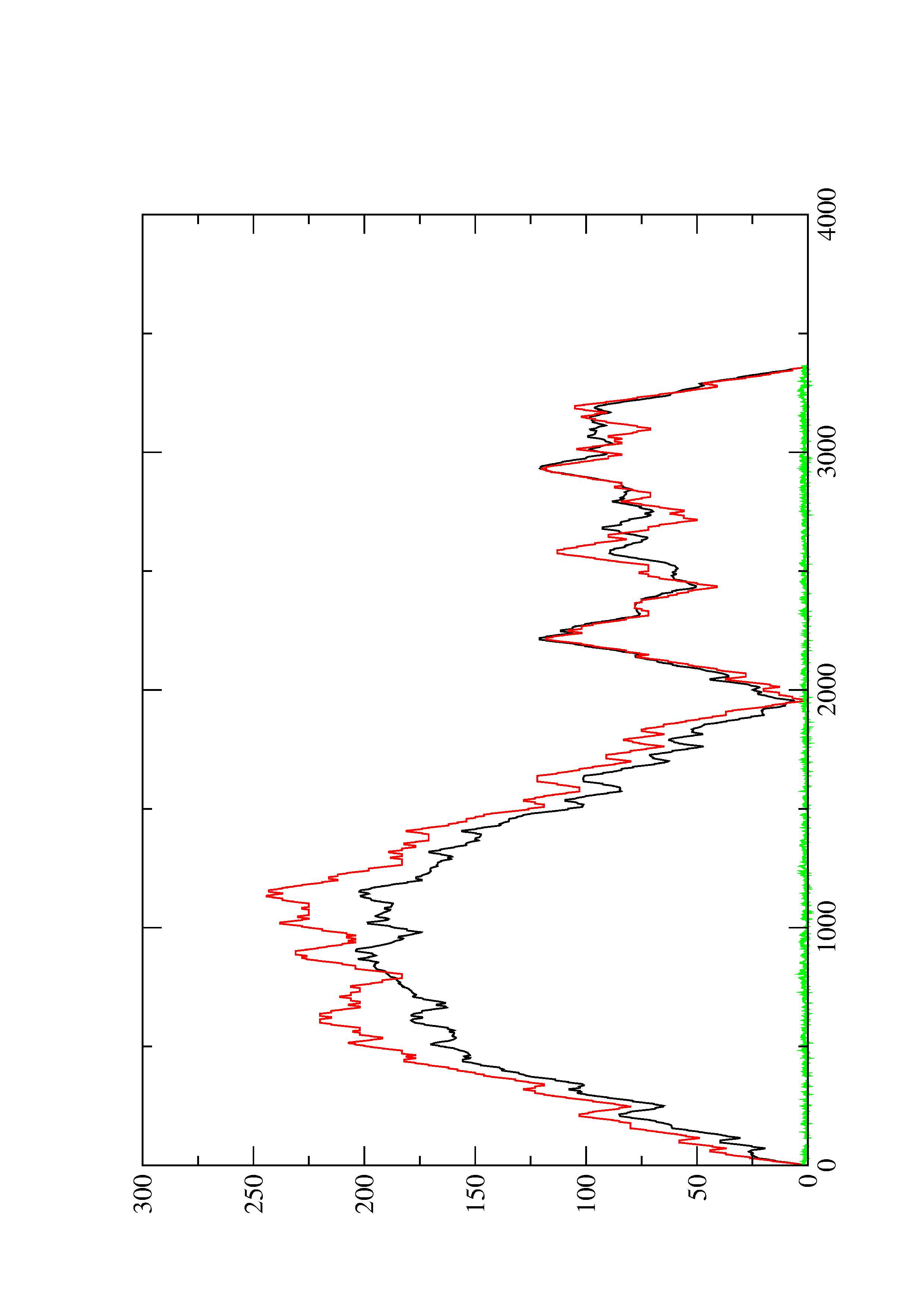
1. The free energy of the thermodynamic ensemble is [-703.53] kcal/mol.
2. The frequency of mfe structure in ensemble 6.92333e-48.
3. The ensemble diversity 861.15.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.