lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01043479.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-467.34) is given below.
1. Sequence-
UUAAUUUUUAAACAUUGACUUUAUAUUUAUAAUAUUAAAUAUAUUAUGACAUAUUUAUAA
UAUUAAAAUAUUUAUUACAUCAUUAUGACGUCAUCCGAGUAGAAGAGGUUUUUACGUCCG
UCUCAACCAGUCGGAGCUGUUGACCCCCGACCCCUAAGCUCAUCCGAGUCUGAAAACAUU
GCAUAUAAUGAACUCUAUAACAUAGCAAACAAGUAAUCAUUAGGCAUAACCUAGAACAAA
ACCACAAUUAAAACAGAAAACAAGAAACAACCAAUGAACUCAAUUUAACAAGUAACAUAU
GCUCAGAUUCUGUUAGGCAGAAUUAAGAAUUAAUAGAACCAACUCUAUCUUAGCUAUCAU
AUCUUGACCAAGUCUUACAUGUAGAGUAGUUAGAGGGAGAUAUACAAGUAUAUUACGAAA
UUUUAAAAGCGCAAACAGAUAGGCAGCAUGAUAGUCAAAGGUUUACUGAGCAGAUCAAAU
UAACACUCAAGAAUAUAGCAGCUAUAUCUAGGCAGAGUAAAGCAGCUAACUCAUAUGAAC
AAAAUACUAAGAUUAAGCUAAACUGAAUACAAACAGACAAGUUAAGACUAAUGUUGGAUG
UCAAUAAGCUAAUAGGCAACAGAGGGGUUACACAAGACCAAGUUAAGCUUUUAUUAGAUA
GUCCAUUAGUCAGCUAGUCAAGAAGAUGCAAGAAAGAAUCACACAAUGUUGUUGAGGCAG
GUGAUCAAACUGGAAAGCAAUAUAUACAAGUCUAUCAAUACUGGAGCGGGGAGAUAUGCC
UAGGGCCCAAUAAAGAGUACUCGGCAUUUUCAUUUACUUAAAAACAAUGGAGAUCAUAUU
UAAAUAUCAUUCAUCCCUCAAACCUUUGGACUGACUGGAGUUAAGUUGGACCCACACCAA
CUUGUGGUCCUCACCUUAGUAUUCUUGCAAAGUAUAAGCCACUAGGAUUGAUAAAAAAAA
GCCUUGGAUUCCCAGAGUUGACCUUACAAGUCUACUCCCUUUACUUGUGCUCAUUAAAGU
CCUUGGGCAAAUUAAGGGUCAUCUUUGAAAAAAACAGAAUGUGGGGUGGGUUGAGGGGUU
UUUGCUUUAUGACUUUGGGGUUUCAAAGUUAUUAUAUAAAAGUCACUUUGAAGAAUAGGG
AUCAAAUGGCACUAGGAUAGCCUCCCUUACCAUACCAAUCCUAUAGGCAACCUGAAUACU
UUAGCCCUUACUUUUAAAAGAAAAAGAACUUUUAUAAAGGAAAAUAUUAGGGCCAAACUA
UGAGUGAAAUCUCCCCUUUAAUGAAAAGUAGGCAACAUUUAAUCAUAGACAAAAACAAAA
AACAUAUAGCAAAUAAGGGACACAUCUCAACUGAUAAAGACUAUUGGAAAGCAUAAUAUC
AUAGAGACAUCAAAACACAUCUUUUUAGCCUUAAAACCCACUCAAAAACAAUACUAGCCU
ACAUAUUGAAGGUCAGACACAUAGCAUUUAGCAAACUAGUAAGUAAACUCAUUCAACUAC
UAAGAAUCAUUAUCAUCAGAGAAAUAAGAACAUAACCUGGUUAUAAUCAGAAAAAGGGAC
AACAUUUUGACUCUAGGCUAACAUAGAGCCAACAUUAUCUUUCAAUUCUAACUAAGUUCA
AACAGUCAGUCCCAACAAACAAAGUCAGUUUCAUGUAACAAUUAAGAACCAGAACCAGCC
AUCAUGAUCAGAAAAAGACCAUGCUUUUAUUUAAGCUAUCACAUGGAAAUAUUAACAACU
AGACAAGUUCAGUCUAGUUCAUUCUAGGCAUGCAUAUUUACUUUUCAAUUAUAUUAGAAC
ACAAUCAAUGUAUCAAAAUGACCAGAAAUAGAAUCCUACUUACACAGGAAAAUAGCAACA
CCAAAUCAAAUCUCAUAACGUCAAAACCACCUCCUGAACCCUCUAUAAAUCAAAGUUAAA
GCUAAAUCAAAUCAAGCUACACUAACAACUAAGUCCAUCCUGCCAUGAUGAUAAGAAAAA
GCAGCACACCAUAUUUUGAUUAUCAACACCUCAAGUAGAGUUAACUAAAACAGUAAUAAA
CAAAGCAGUAUCCAGCAGUGGCUAUAAACAUGCAAAAGAAUAGCUUAUGCCAGCAAAUAA
UGUAAUUGAUAGAGAAAAUAAAGGCCAAGGAGUAGUAUAGUACCUUCUUUAGAGUAGGAA
CCAAAGUUGAACCACAUAAAUAACCCAGAAAUCUUAAAAUAAACUAAAAAAACAGACACA
AAGAUCUACAAAGCCUACCAAACCCCUUGAAGGAGUGCAAGGAAAUAUCUUUUGAAUUAA
ACACUCCAAUGAUCGAGGAUCACUUUGAUUUUACCUCUCUUAGUACUGAAUUAGACUUAG
AUUAUGAACUUAAUGAACUAAGUAAAAGAUUCACAGUAAGGAUUCCAGCCUAGAACCACC
UUACUAAACCAUAUUAUCAACAAUACUUACUAUUUUAAUUUUAAAACUAGCAAGCUUAUG
CCAAAAAUUAACUAAGAAAUUAGCAGAACCUUAAACCCUAAAAAUGCUCAUUGUAAAUCG
CUCAAUGAAUGAAUAUCAAUCUCUCUCUCUAUUUAUGAAUCAAUCCCCUUGAACCCAAAA
AAGGCAAAGGAAAUUUAUAGGAAACCACAACUUGUUCUCCAAGACAAAAAAUAAAGGGAA
AAACAAUACUCAACUUGGAAGAGAAAAAGUAAAGGGAAAAACAAUACUCAACUUGGCAUA
GAUGAAUCAAAGCCCUUAAUUAUUAAAAAG
2. MFE structure-
...............((((.(((((.....((((((....(((((((((.....)))))))))...))))))...........))))).))))....((((((((.((((((...((.(((((...((((...(((.(((((...(((..........(((.(((((((.(((....(((((((.(((((((((((((..
.))))......)))..))))))(((((((.(((..........................(((.(((((......................((((.....))))..((((((((...))))))))........(((((......)))))..((((((((((((...(((...)))....)))).)).))))))((((((((
(((..((((((............(((((((...(((.((((..((((((..(((((..(((((...(((.....)))....))).))........((((.(((...(((....)))....))).))))...........................((((...(((........)))...))))..)))))))))))..))
)).....(((....)))........((((......))))..)))..)))))))...(((..((((((.((.....(((.((((((((((..((.(((........((((((((....(((.........)))...)))))))).......))).)).....(((.(((((........)))...)))))...........
...)))))))).)).))).....)))))))).)))))))))..))))).....))))))........((((((((((.....(((((((.......)))))))(((((......(((.(((....))).))).....)))))(((((((((.............(((....)))((((.(((.......))).))))(((
(((....((((((..........))))))...))))))...(((((((..........((((((((((((((..((((((((((((.((((..((((...(((((((((.(((.......))).))))))))).((((((.....((((....(((........)))..)))).....))))))(((...))).......
...(((((..((((((((((........)))))).)))).........)))))....((((((.((((.....((((((.....))))..)).....)))).)))))).......))))...)))).))))))...((((....))))..((((((.....))))))...............(((((............)
))))..))))))..)))))))))..........((.((((.((...)).)))).)).))))).((.....))....(((((.((...........))))))).............)))))))..............(((((....)))))....((((.(((....))).)))).((((.......))))..........
.)))))))))............))))))))))...............))))).)))....................((((((.((((((((((........(((((((.(((....((((((((((..........((((((((.......)))))))).....((((.((...(((((.((..(((..((((((.((..
....))))))))......)))..)))))))..))))))......(((((..........................................))))).................(((...((((...........))))..)))...................))))).)))))))))))))))..........)))))))
)))......(((.....)))...(((.....)))..........((........)).)))))).................))).)))))))........))))))).((((((((.......(((....((((....((((.......(((((((((........(((((....................(((((.....
....................)))))...................((((((.((((((.(((((....))))).........)))))).....))))))...(((..((.........))..)))..((((((..((((((.((((.......)))).))))))....)))))).(((((((.(((.......)))...))
)))))...........)))))........))))))))).......))))....))))..))).........((((....))))........................((((((.........)))))).................))))))))(((((((...(((((((..........))))....))).)))))))(
(...)).....(((((.(((................)))..)))))...)))))))))).)))..........))).....))))))))...))))...))))).)).)))))))))..)))))......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-518.44] kcal/mol.
2. The frequency of mfe structure in ensemble 9.75589e-37.
3. The ensemble diversity 754.89.
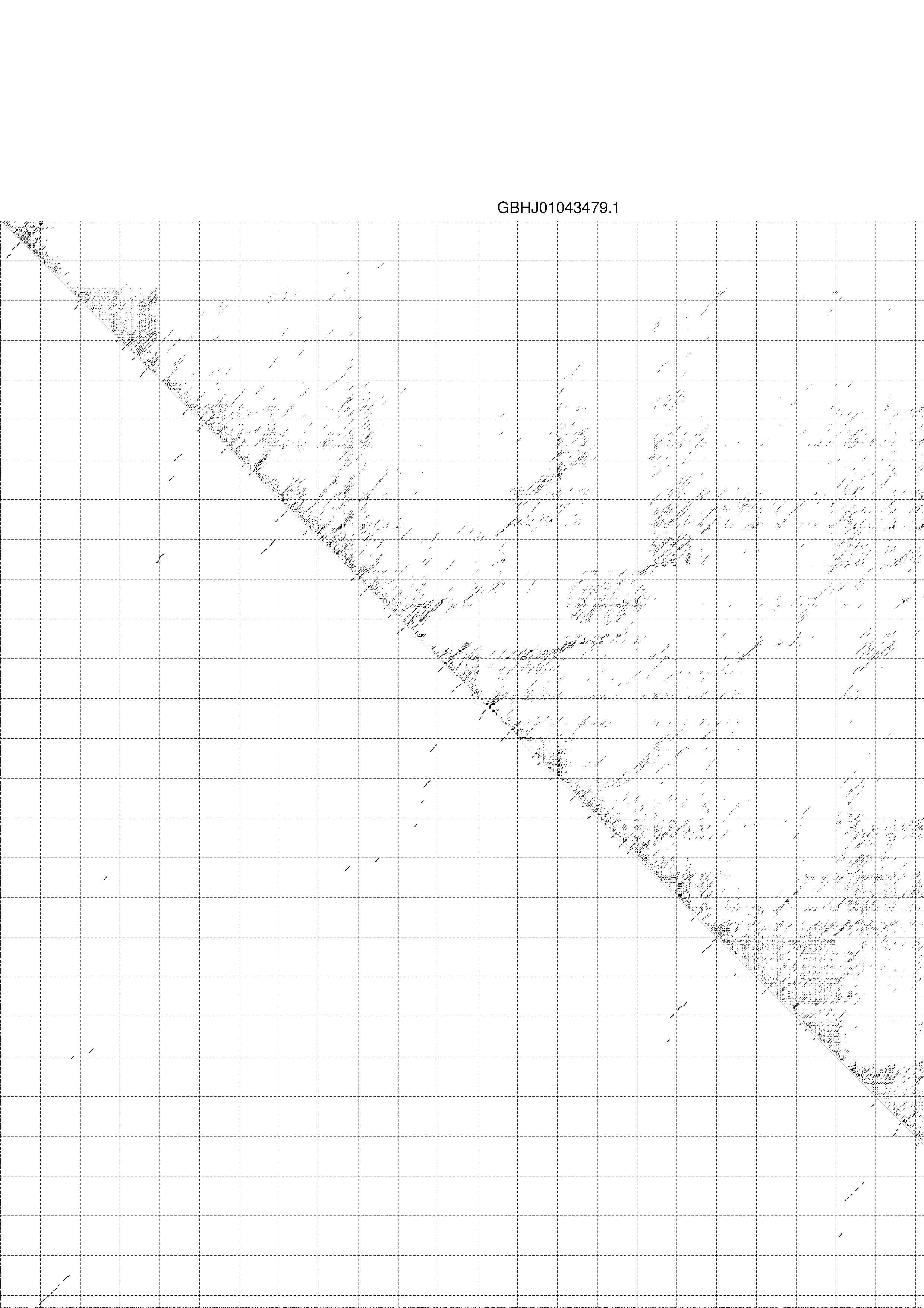
4. You may look at the dot plot containing the base pair probabilities [below]
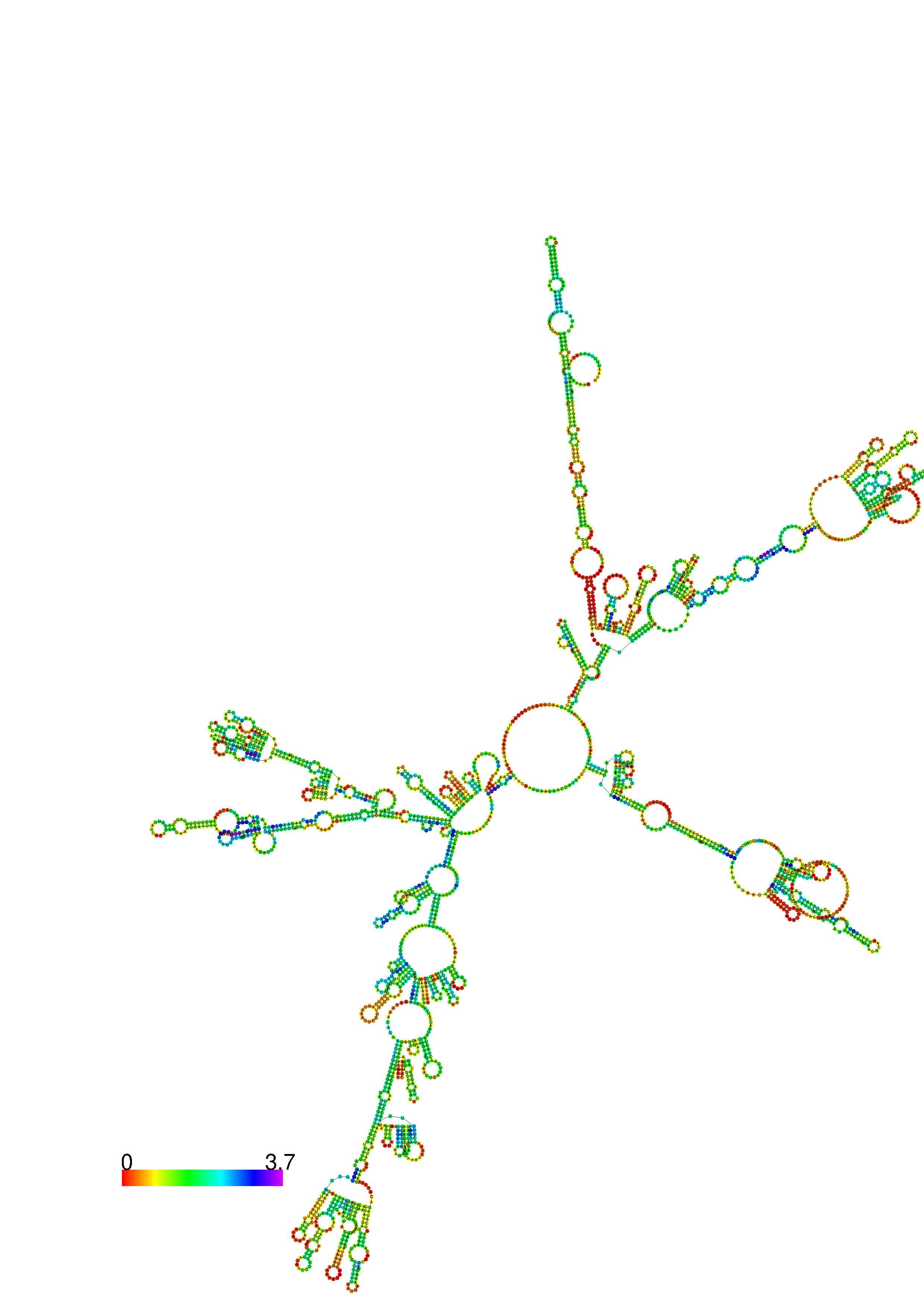
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
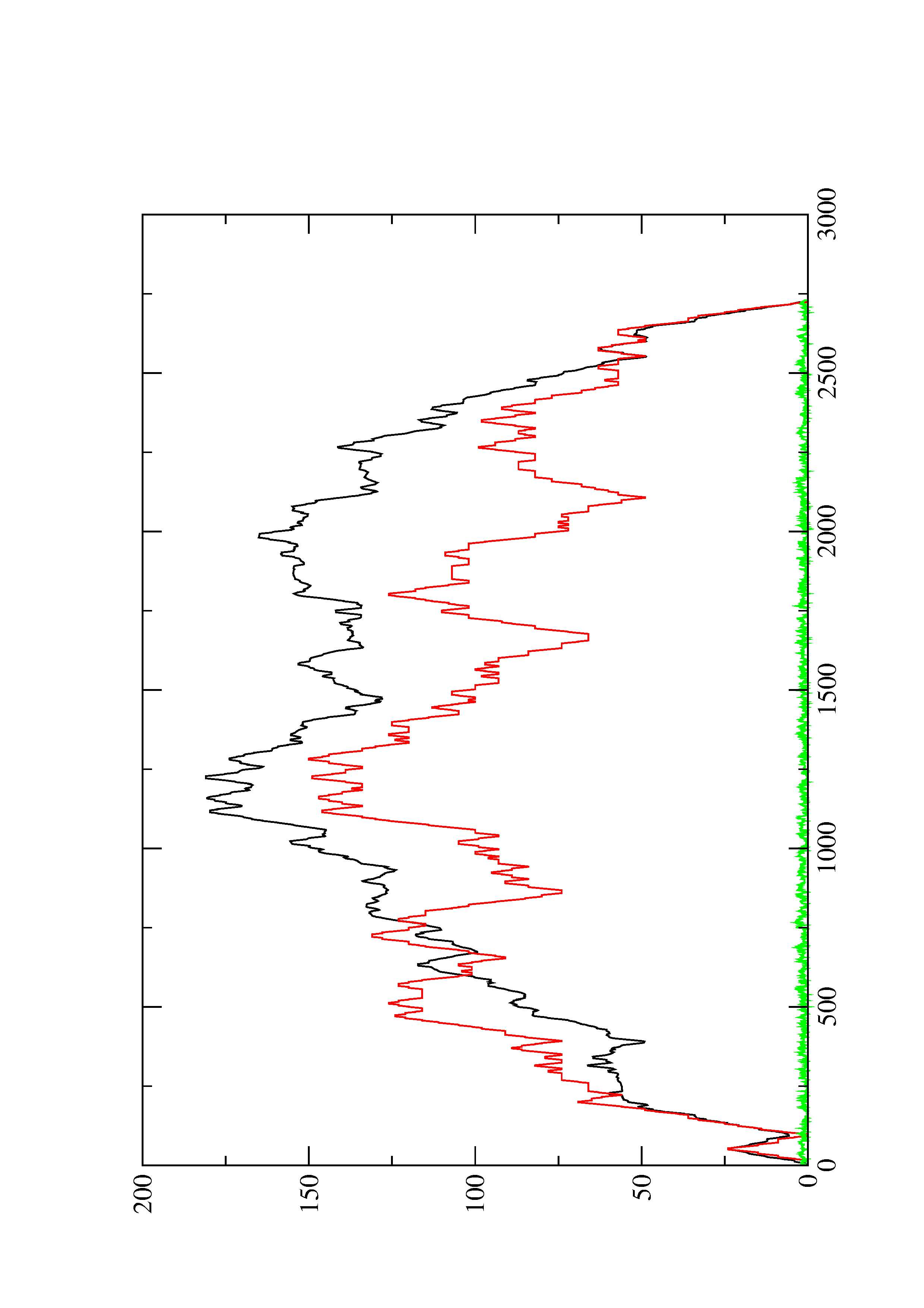
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.