lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01043866.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-477.29) is given below.
1. Sequence-
UAAACGUCAAACUUUUUAUUAGCUAAUUAAAUUUUUUAUUCAUCUUUUUUUUUUAAUUAC
AUAAAUUUUUUUAGUGAUACAUUUUAUUCUAUUUGAUACAUUAUAAAAUUUAAAAUAUUA
AAAUAAUUCAUAAUACAUUGCAGUAUUCAUACUUUAUACUACAUUGUCGUAUAUAUGAUG
UAUGAUAGAUCUUCAUGAUGAAUUUUAUUAUACAUUGUAAUAUUCCAUGGUACAUUACAG
UCUUCUUCUGUAUCAUCAUCCUCUUCCUCCUUUUCUUCUUCAAAUAGCCUAACCGAAAUU
CAAAAAUCAACUAAAUUUUUGGGAAAUUGGAAUUGAAAAAACGGUGGAAUCCUUUUAAAU
GGCUGGCAAGUGAAAUUUGUAUGAUAUUAAAUGAAAGUUAAUGGAAGAUGGUGUGAUUAU
UGUUGUGCAAAAUGGAAGUGUGGGAGAUUGAAAAUUCUUUUUAUUGGAAAAAAUCUUUUC
UUCUUAUACAAUGUGGUUGUUUUUUGUGUUUGUGUGUUGGUAUCAAAAGAGGGAAAGGGA
AGAGAGAUAAAGAAAAAUAAAUAAAUAAAUAUUAAAUAUAAUAAAGUAAAAGGUGGAUAA
AGAAGAUAGUAUUAAUAACAAAAUUAGAGUAAUUUAUAUAAUUUUUUCAUAACUUCAGUC
UAUGAGAACUAACUAAACUAGAAGCGUCAUAUUAACCAAUUUCUUUGGAUGUGACAGUAC
AAAAUAAGCCUAAUGCCUCUUUACCCCUCAAAGUUAAAGGGAUAUAAUUUAUGUACAUAU
GUCCCUUUUUUAUGAGUUUAAAAUUUAUAUAUUGAUAUUCUAAAAAGGAAUUUUAUAAUA
UCAAAUUAUUCAGAAAAGUAAGUUGAAAUAACUUUUCAUAAAAAGUGAAAUUAAUAACUU
GAAAAGUGAGCAUGUUAACGUCAUUAGCUGUUAAAAUUUUGUUAAAUUACAGUGACAGAA
UUAAAAUUAUUUUACACUAAUAUGUACUAUAAGAGUCGCAUUCUCCUCAACGAUCAUGCA
UAAGUUUGGGACGCCAAACUCAUUUUGUACCCUUUUUGCAUAUCAAUGAUCAAAUAGCUG
UCUUUUAAAAUGUACUAUAAGUGAAAUCCUUCAUUAGGUGUCAAUCUUCCUUGUACAAAU
UGGUAGCAAGGGAAUUGACAACUUCAAGAAGAUGUCGAAAAGUCAAAUAUAGUUACCAAU
GUUUGAAAAAUAUUUGUCAGCAUAUCAUGGUCUUUUAAAGAAUAUUUCAUAGCCACUACG
UGUGUGUGAAUCAUCCAAAAAGGGACUGUACAACAACCUUUUUCGUUUUUUAAUUGGACG
UGAAAACUCAUUUUUCUUUUUAUCGGUAUCUAAAUUGGAACUGUAAUAUAUAUUAUGUUA
UAUCUAUGCAAUGCCUUUAGAUCUUUUUUUUUUUAAAAAAAAACAUUUAAAGAACAAAAU
UUCCAUUUGACAUUGAAUAUUGAGCCCUACAUAAGCACAAUUUUUUAUUAACAUAGCAAG
AAAAUUUGCAAUUGCUAUUCUUUAUGUGCAUUAAAAAAUUCACUCUUGUAUAAUCACGAA
CCAUUCAUGAUAUAAAACCACAUUAUACAAGCCCAAUACGAUGAAUUCGACAAGAGAAUC
AAUCUUUGACAUCUCUUGUAGGAUACCACAUGUCACAUCAACUCAACCACCUUAACAAGU
AACAUAAGCAUAGGUAACAAGCAGUUCGAACUAUUUGAAAUUUUCCACACCAAAAAUAUU
AGUUUAAAUGAAAGUAUUUGUGCUUGAAUAAUUUUUUGAAUCCAUAAUGCAAUCAAACAA
AUGAGAUGAGCAAAUACACUUCUUGAUACCGUAUUAUCUGUCUAACUACAUAAAAUUUGA
UAAAUGCUACAAAAUGUGUCCAAAAGAUAAGAGGGCACAAUCUUCACUUAGCAAACUAUU
AUAUAAAUACACAUGAAACUUUGGAAAAGAAAAAAAAUUAUAUCAAAAAGCAAACAACCG
AUGGAUGGUGUCCAACAUUUUCUAUUAAAUAUUAAUGAAAGUCUAUACAUACUGUAUUCA
CAUGGUAAAACCAGCUAACUCCUAACAUUAUUCCAACAACGAUAAAAAUAUAUAGCAAAU
AACUAAUUAAGAAACAAAACAAAACAAAACAAACUCUAACGCUGUACAAGGAACAAAAAC
AGUAUUAACUUGGUGCAAGUUUAAUGUUUUCAAAGCGAUUGAAAGGGUCUCAACUUCAGG
AAACACGCGUGAAGCAUCUCUGUAAAGCCCAAGACAGGAACAAAACAGGACUACAUGAUA
UAUCUGGGAAUGAUGGUGGUCGCUGAAGGAUAGAAUUGACGAGCUCCUCGUUUAGCCUUA
CAGGGAGUGGAGGCAUUUCAAUCAUCUCUAACGCGUCAUUCGCGCUGCAAGAGAUAGCAC
GUUAUGAGGUGGAAAGCAAGUCUAGAGAAUGACAAGAAGCAAAAAUAAAUACAUGCCUUA
AAAAGAUAUGUAAAUAUGGGUGGUGGGUG
2. MFE structure-
.....((((..((((..(((.(((.((((..(((((((((((((((((.......(((((.((((....)))))))))..((((((((.(((((((((..((((...(((((......................(((.((.(((((.........))))).)).))).((((.((((((((((((((((..((((...))
)).))))))))))))))))))))...((((((((....((....))..)))))))).((((((((((..(((((((((((......(((.(((((((((.((((((.((((((......(((((....((((..((.....((((((((...((((.(((..((.(((.......(((((((((((.(((......(((.
......))).......))).))))))))))).......))).))..))).))))....)))))))).......))..))))))))).......)))))).)))))).)))).)).)))))).....)))))))))))))))))).))).........)))))..)))).))))))))).)))))))))))))))))))))
))))..)))).............((((((..............((((((((((....)).))))))))...............(((((((((...((((.....))))))))))).)).............(((((((((..((((((..(((..(((((((((((...........)))))))))))..((((((((..
.))))))))..((((((((.(((((.....))))).))))))))........(((((((...........))))))).....((((..........))))((((((((((....((((((........))))))((((((((((........))))))))))............((((.(((.((((..((((((..((.
......(((..(((.(((((..(((((((....)))))))................))))))))..))).......))...))))))....))))))).)))).....((((...((((((((((.((((((((((......)).)))))))))))))).))))...)))).((.((((...((((((((........))
))))))......)))).)).....(((((.((..(((((((((..((((((..(((...)))..))))))......(((((((.............))))))).)))))))))..)).)))))...)))))))))).((((((((((((......((((..(((((((((...)))))))))))))....)))))(((((
((.((((((((....)))))))).)))))))........(((((.........((((..((((((((((((..((...(((((((((...(((((..(((((.....((....))..))))))))))....)))))))))..))..))))..((((.(((...))).))))........((((((.((..((((((..((
(.....)))((((((((.((((...))))..)))))))).((((((((....((.((((...........................((((((((((.....((....((((((..........................))))))...))....))))))))))(((((((((((((...((((.......((((((...
(((((.((.((((((((.............)))))...))).)).)).)))....))))))...(((((.....(((((((...........))))))).........)))))............................))))......)))))))))).)))..............))))))))))))))...((((
((.(((((...)))))))))))......(((((((.......((((...))))......((((....(((((........))))).....((((((............................................))))))..))))......)))))))....)))).))..)).))))))((((((.....))
))))))).))))).)))))))))...((.....)).((.((((..........))))))......(((((((((......((((.......)))))))))).)))...)))))))..((((((...))))))....)))..)))).)))))))))))........))))))(((.((((..(((.(((........))))
)).)))).)))...))).)))..))))..))))......((...(((..((((((.(((...))).))))))..)))...)).......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-526.17] kcal/mol.
2. The frequency of mfe structure in ensemble 3.57681e-35.
3. The ensemble diversity 644.37.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
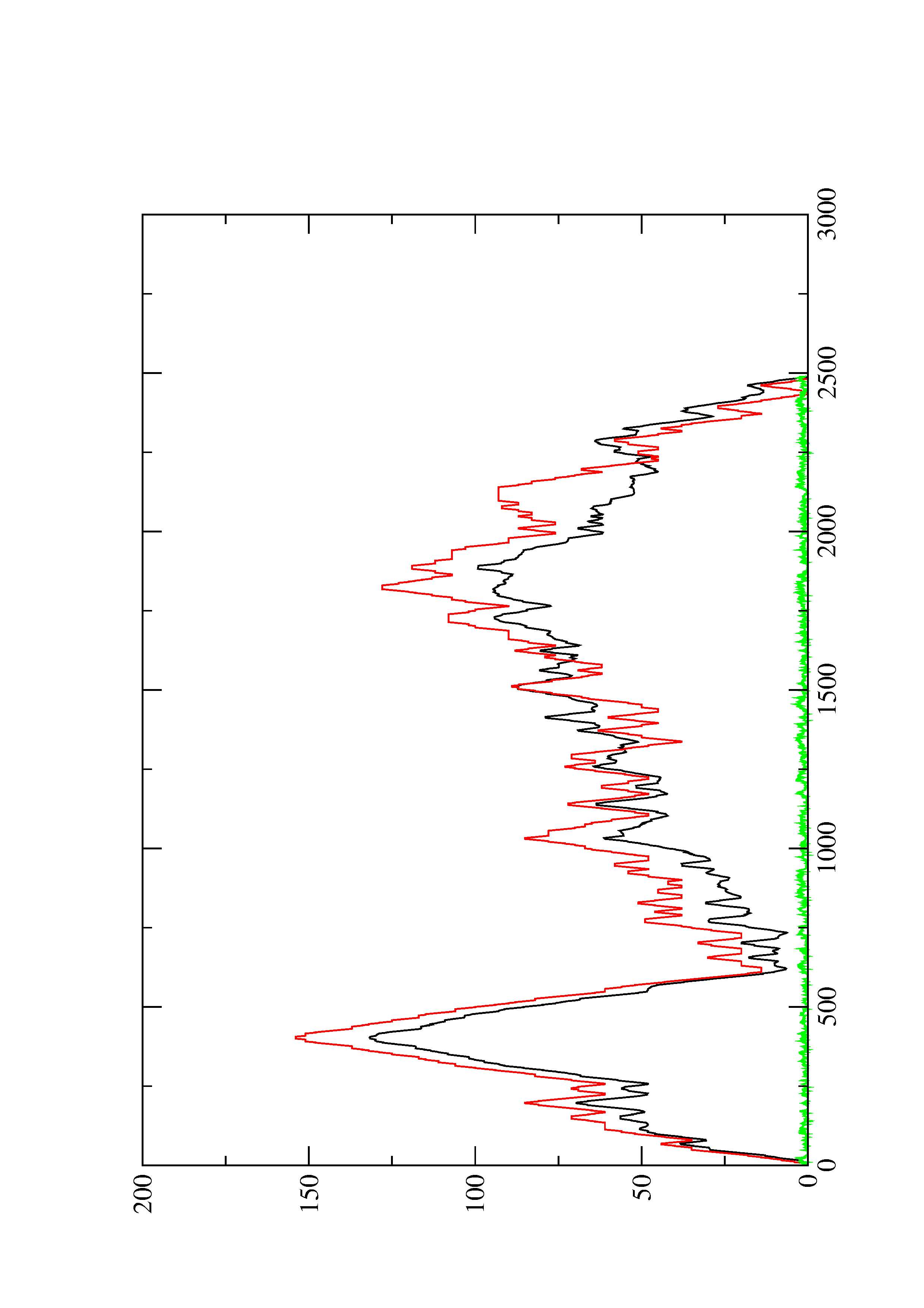
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.