lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01043981.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-560.20) is given below.
1. Sequence-
GCACUGGCUGCCAGGAAUCCGUUGUCAGGUCUUCCCUCUGCAAAUGGAAGCAUGCUGCCA
AGCUCAUAUCCUGUAUGAACCUAUGUAUCAUUUAUCUAACUGGCUUUAUGUCUUGAAUCA
UUACUAGUAGAAUGCUAGUCUUAUCUUGAAUUGGAUUACUGAACUAGGCAUUCUUUUGUG
ACAUUCAAGAUCAUGAGCUCAGCUCCAGCCUCGUAGUUGUUGUCUUCUUGGGGAGCAAUA
CAGAAAGUUUUUAACUUAAAUUAGCAAAUCAGUUUUUAUGAAAGAUAUUUUGUGGCUCUG
AUCCUCUCUGACGUACUUCAUAGUGUUUUUUGUCAGGCCAAAUUUUGUUGCCUAUAUUUU
CUCUGUAUGCUGGAAUUUAGCACUGCUUAUAUUUAAAAUAUUGGAUUGCUGAUGGAAGUU
AAAUUACUAUGGCAAUGUUUGACCAUGUAUAAGAAUAUCUUGGUUGCUAAUGUUCAUUGA
AAUUUUCUUUUGUCCGUGUUGUUUUGCUAUAAUCAGUGUGAUUUUUCAUUGCUUUUUCUA
UUUCCCCUGCAACAUAGCAUCGUUUCCAUUCCUUUUUCGGGUAAUGAAAUUACAUACAGU
UUUAAUGCAUUGGAAACUUCCAAGUUGUUUAUUCAUAUGUCAAUAUUUCUACAUUGCUUG
UCCACAAAGUCCAAAUUAGUUAAUUCUCACAAUUUAUGCCUUUCUCAUAACGCCAUCUUU
UCUUAGCUACUGUGGAUUUGUCUGAUUUCCCUUAUGGCCGAGAUGAAGGAGAAGGGAAGA
AAUAUUUGAAAAUGGAUCAAAUUUAUGUGAAUAUCUUAACAUUGUUCUAUCUGGUGUUUU
ACAGGAUCAACCUCCUGUGAGUGUAGCCAGUCUAUUAAAUUCGCUCGGUUUGGGGAAGUA
CGCUAUUUCACUUUCAAGCUGAGGCGGUGACCAGAACUAUUGUUAAUUGAUGAAUGAUGU
GUUUUUAGUUUUUUUAUCGUUUCUUUGUUAUUAUGUUAUCUCAGUAUUACUGUGGCAGAA
AAAGUUGGAAAGGUAUACUGGAUUUCACUGCAAAAUUAACAAGGUCAUGAGUUAAGUAGU
UUCAUGUUGGAUAGACAACCUUCUUGUCUUCUUUAUAAAAAAGGAUAUUUUUUAAUGAGG
AGCAAGUUAGAAUGGUACUUAAUAUUCUGCGUCAAAUCUUUUCGCCUAGCGUACAUGAUU
CCUAAUUGCCCAAAUCAAUCUUAAAAUUUUCUUCCUUUGUUUGAAUCACAUAUCAAUUAC
UAGGUCACAUGACCUUUCCUACUCCGUUUGAUCUACCUCUUUGUCUGUCCUCCCAACUAA
UGAAACUUGUUAAAUAGAUGCUUUAUGGGCCUUCUUUACAUAAUUGUCUUUUGUUGUUUA
UUAUACUUCCGGAGAGUUUUUUUUUUCUUGAAACUGGUAACUUACUUCCUGAGAGGUAUU
UGUGAUCACAUCGAAGCUCUUCAUCUCUUUAUUGUCCUCCCUUUCUAACCAUAUGGUCAU
AUUGGACUUCUAAUCCGAAAUUAUUAUGUGUCAAUAAUAGUAACCUCACUGCUAUUCACA
AAUUAAAAAGUUGCACCAUUGACCCAUAGCAUUUUCAAUCAUCCUCUAAUUCGCCAGCCA
UGUCCUUUGUUCCACGGCUCGACUGAAUGAUCAGUCCAUCCCUUUUAUAUGUUUGUUCAC
AUUCCCCAGUUCAUGUUUUCGCUUGAGUAAUUUUAGUAUACUUUGUCAGAAUUUGAGAUA
UCCUAGUUCAACUCCUCCUUUAGUGAUGUGACGCCUUACCCAUUUCAGGUAGACAUGGCU
ACAUUAAAGCAAAUGGGGGAUCGUGACCUCAAGGAACUUGAGAUACCAAUGGGUACUAUC
AUCUUUGUUGGACAAUCACACUGUUCACAAAUUCUACUGCAAGUUAAUAUUCACUUCCCG
AAGUUUAAAGGAUAAUUCUCGUUGGGGGUGGGAAGAGGGCUAAACAACUCUUGAUUACAC
UGAACUGCUGUAGGACAUUUGCUGCUCUUUUUCCAUUUUAGCGUCUAAUUGCUGCUCUCU
GGGGAGUUGCAGGGACCAAGGAAGAAGAUUCAGGCUAUGCCUUCCCAAACUAGAAGGAUA
GUGUAGUAUGGUAGUUGUGUUUGUUGAACGCAUGCUCGUCUGUCGUUAUGAAGCUGAAGU
UUUCAAACUGGCUGUUCUAGGGAAAUGUAUAUGAAGGGUAGUGGUAAAAUGCUGGAGUAU
UUGAAGAAUAUUUGUUGUAUAAAAUAACAAAAUAGCAGUGAAAUUCUCUCUUUGUUAUUG
GGUUGAGGCUGACAUUUUUGUGAUUUUAUUUUAGAGAGGAACUGUAACUGUUCGACAUGG
GAAGAGGUUUGGAGUUAUUCCAAUAGAAAAAAUU
2. MFE structure-
(((.((((.(((((((.....((((((((((...((.........))((((((..(((((...((((((...)))))).....................(((((((.(((((((((((..((((....((((((((((((..((((......))))....)).))).)))))))...)))).)))))))).))))))).)
)).......((.(((.(((((.(((....))).))))))))))..(((.((((((((..(((((((((((((...((((((((....))))))))..)))))....(((((((((((....)))).....))))))).............((.(((((((....((((((((.....))))).)))........))))))
).)).))))))))..))))))))..))).))))))))))))))).)).)))).....))))))).)))).)))((((((.......(((.((.((.(((((((((((...((((((((((.(((((((..(((((((((((((((((...(((..(((...(((((((..((......))(((.(((((((......)))
)))).)))..)))))))...(((((((..(((((((((((((((((((((...((.(((.(((...(((((((...(((((((..........(((((.......)))))............)))))))...))))))).....(((((((....))..)))))...))).))).))))))))).))))...........
...))))))))))...)))).)))......(((((..(((((((((......)))))))))....)))))............)))..))).))))))))).(((............))).((((((((((..(((((((..........................))))))).))))))))))..........(((((..
.((((...)))))))))))))))))))))))).)))))).)))).....)))))).))))).)).)).)))((((..(((((((((((((((((((((....((((.((((((..(((((((.....)))))))..)))))))))).(((((((........))))))).....((((....(((...))).....))))
..........((((.(((..................))))))).((((.............(((((....))))).............))))........))))))))))....)))..))))))))..))))..(((((..(((((...........))))).)))))((((.(((((((.((((((((((.((((...
.....((........))......)))).))).)))))))..))))..))).))))(((((..((((((((..((((....((((((((..(((.(((((((((((.((((.(((((((.((((((((......))))))))....(((((((((((..(((((.....(((...(((((((.(((....(((...((...
.(((.(((......((((((.(((((((.((((((((...((((((....))))))...))...............((((((((((((.((((.(((((.((..(((((.........))))).)).))))).))))(((((((((.............)))).)))))...((((((((.......)))))....))).
.............)))))))).)))).((((((...))))))...((((((((...((((...(((((.(((((.......))))))))))............((((..(((.......)))..))))..))))...))))))))..)))))))))))))...((((((((..(((((((((..((.(((((((.(((..
.(((.(((((((((........((((..((((.(((((....))))).))))))))...)))))))))..)))...))))))......)).)).))..)))))))))..)).))))))(((((..((((....((((((........))).)))....)))).)))))))))))...))))))..))...)))..)))))
))))).....))).....)))))........(((((((.....))))))))))))))))))................)))))))))))))))......))))))).))).))))))))((((.......))))))))...))))))))..))))).....)))).)).......
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-608.15] kcal/mol.
2. The frequency of mfe structure in ensemble 1.63839e-34.
3. The ensemble diversity 659.32.
4. You may look at the dot plot containing the base pair probabilities [below]

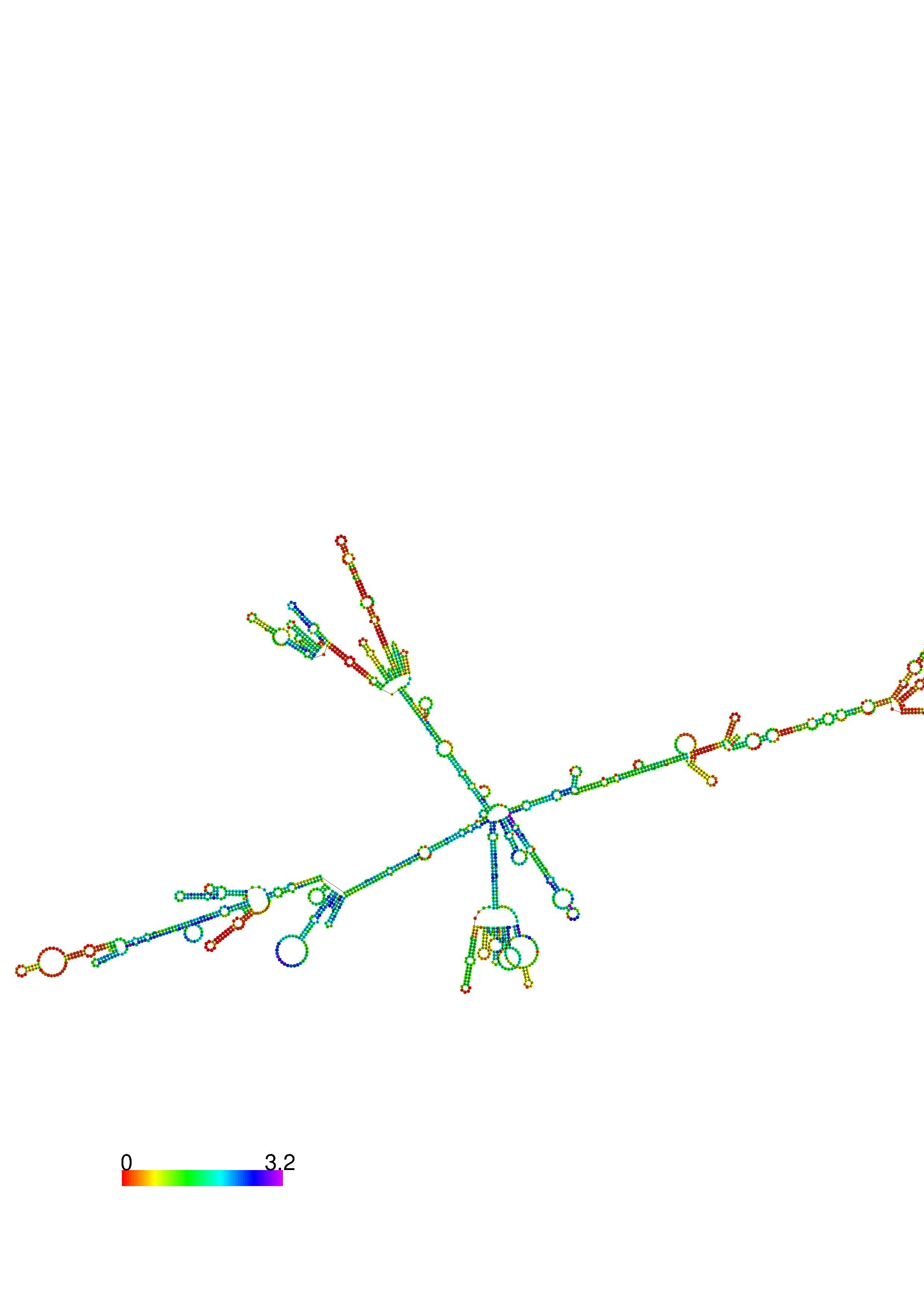
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
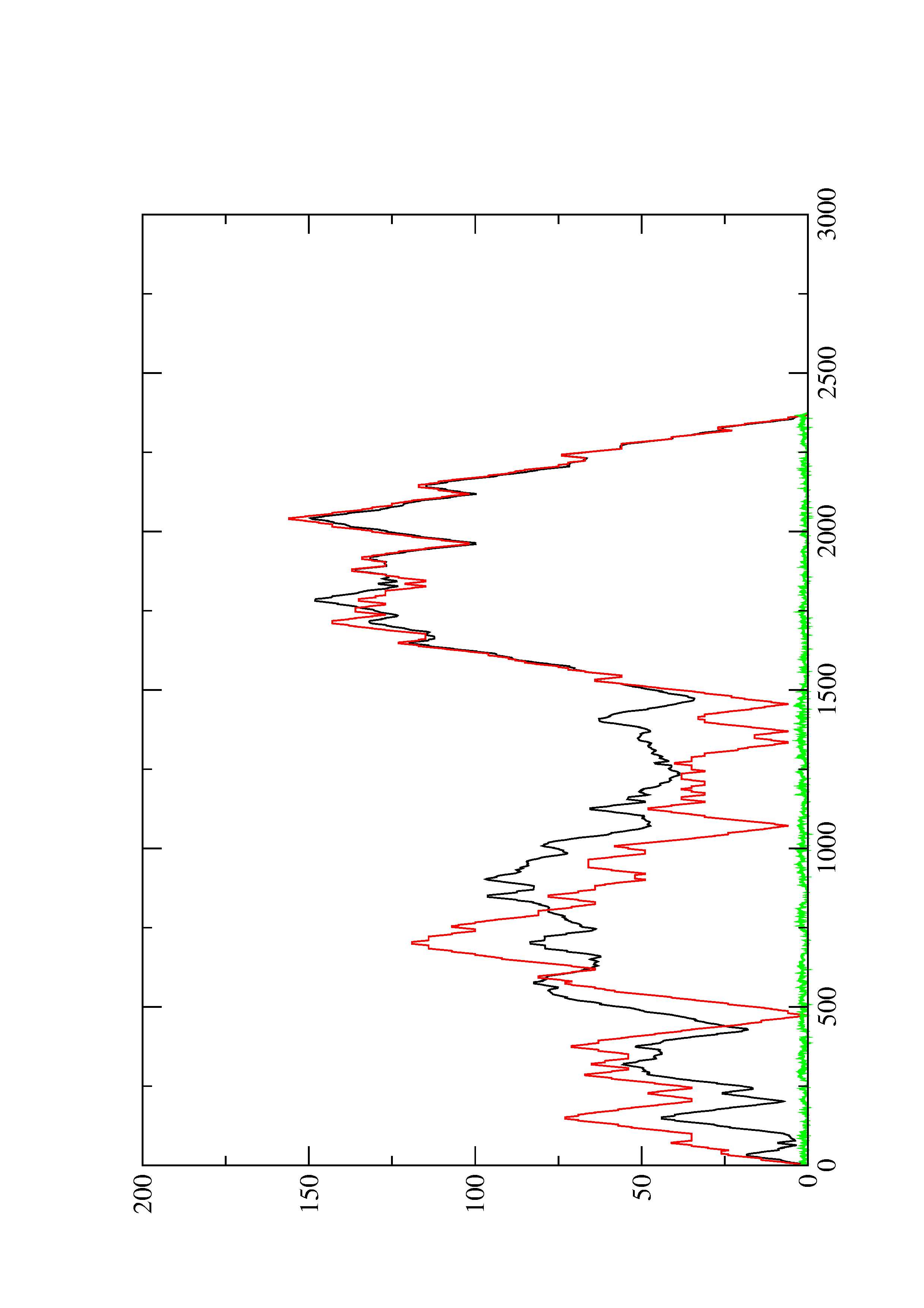
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.