lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01044128.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-470.60) is given below.
1. Sequence-
AAACAGUUCAGCUAUUUCACACAUUAAAGAUGGCAAAAUGAACAAACAAAGAGAAAGAUA
AGUAAUAAAAUCUACAACAUAGUAGAAGCUUGAUUCAACAUUCCUCUCUAAAGAACUUUC
AUGCUGCAUGAUAGCAGCAUAAGAAUUUGCAUAAGAAAAGCCUCCAGUUGUUCUACACAU
UGCUUGCAAACUUGUGCUUCACAUUGUAAGAAAUCCGCUUUGAGCCGGAAAUUUUCUUCC
AACAGUCAUAAGCUAUACCUCACUCAAUCAAGCAUGUCCACCACAAAUGUAAUCAUUCUU
GGUAACAUUGGUCACUAUGUUAAACAGUUACCUGCUCAAUCUCAAUGGGAAGAUGAGUUC
CAUUCUUUCAGCAUGCAUCUUAGUCCAUUGCUUUGACCUCAAGAAGUUAAAAUCGUCGAG
CAUUGUCAGGAACCAUGAACUUCUCUAUCAUGAGAAAGCCAGUUGAGCUUGCACACUAGC
AUUGAUGAAAAACACCUGAAUCAAUAAACUUUAGCUAGAGGGAACAACUUCAAGAUUAUA
CAUUCCCAAAGGUCAUAUUUACAUGACUAAGCCAAGAACAGCUUGGUUCAAUCCAUUGGU
CAAGAAACUUACCAUGAUGGCGCCGGCACAACAAUAUAUCCAUAGAGGGAUAGGUAAGUA
CAUAGCAGGAACACUGCCAUGGAUGCAAACCAUGAUAGCAUACAACUAUAACUCAACACU
AAACCACGUGGGUUCAACUGUAUAAAUCAUUAAUAUCGACAUCACUCCAUUUGGACCUGU
UACAAUCUAAUACCUAUAAAUCUAUGCUUAAAAAUUUAUAAUACUUCCUGGUAAAUGGUU
CCACAUGCACCACCAAGAUUCAGGUCCACUCGAGUUAACCUAAAUGCAUUAAAAUCAAGA
ACAUUAGCAUCACAUCCUACACAUAAGCAUCAAUAAUGCUAUUGUAAGUGAUAUUUUGUA
CAGGGAAAGAACUAUGACCAAGAUAACACAGUUUUAAAUAAUGGUUCUUGCAACGGAUAG
CAGAUCAUUGAGAAAUCAUUGGCAUUCAGAUGGGAGAAAAUUAUAGUUAGAGACUACCAU
CCCAAUGCUGCCUCAUGCUUGCUUAUUUUUAACUCUGAUACACUACUCUUGAUCGAUUAC
AUACAUAGGCUCAGAUCUAUCCGGAAAUCGGGUAAAAUAAAGGGACAUCAGUCACGGCUG
CAAACUAGAACAGCAUACAACAUCACCAACAAUCUAGUUGGGUAAGCUAAAUGAUGCCUA
ACAUCCAUUAACUCAAUUAGGACCUGUUUCAUCCCAAUACUCAGUUACUCCACUUACAAC
CUAAGUACCUAAUAUCCAUUCCACUCAAUUAGGACCUGCCCAGUAUAGUUACUGACAACC
UAAGUUUUGCACACUUUUCACAUUUCAUGUUGCACCUGUGUUAGCUUCUCCAAAAAUAUA
CUACUUUUGGAGAAUCCAAGAAGCAACAUUUUUGAAGUGUCCAAGCAACAUAGCUUACAA
CACAUUUUGGUUUGCCCUGCCAAGGUACAGUUCCAAUUGACCAAAACCCAAUUAACUCAA
CAAGCUUAUAUCAUCACAGCCUACAGGUUCAGCAAUGAUGCUCACAAUGUUUCUGGUUUG
UGAAAGGGAAAUGCUCACUCCACUUCAAUUUGUUUGCCUUGUUUUUACUUAGCAAAGAGU
GUAAGAAAGUAGAGAAAAUUAAUUGAAUCUUGUGGCCCUAAACUAAAUAAUUAGAAUGUA
CUAUAAUCUCCUUGUGGUCUUAAACAUCUCAUGUGAAAAAUUACAAGUAAAAACAUUCUU
ACAAACACAUUGAAAUAAAGACAGUAAUAUAGUGUCUAGGAAAGCAGAAGGAAAGAUUGC
AAGAAACACAACACAACAUGAAUAUCCAAAAUUCACCAAUAAUUUUAACUUAAAAGAAUA
ACUUACUUCAAACAAAACUACUGACCUUCAUUUCAAAAAUUAGUUUUCUAUUAUCCAGUC
AUCUAAAUGACUAGGAAAAAAGGCACUGUCUAUAGAUUUCUCUAAACAGAAGAGUCAGCA
UAACUAAAAGAAAUGCAAAUAGGACUUCAUUAUAGGAAACUAUCAUUAAACAUAACUAGU
AUUACUCCCUCAGUUUCAUUUUACUAGGUCACUACACAGAAAGUAGAGUCGCGUCAAAUU
UUACUAAUCCAUUUUAGCAAAUCAAAUCAAGACACUCCUAUUACUUUUUCUCCAUACUGC
CUUGGUAUUACGAAUAACUAACUACAUAAAGUAAUACAGACCUCUAAUUAAAGUUUUACA
GAGGCCAAGUAAUAUGGAACGUAAUGAAAUAUUAUUAUUUC
2. MFE structure-
.....((((((((((((..........)))))))....))))).........((((...((((((((...(((((......))))).((((((((.........(((....)))..(((.(((((((......))))))).)))....(((((((....((...(((((((...))).))))...))...)))))))...
..(((((((((((((((((...)).))))...))))))...)))))...................))))))))((((..((((..((((....))))..)))).)))).(((((.((.(((...(((....)))..............((((.((((((..(((((((.....((((((((..(((.(((((((..(((.
..............((.(((((((((...(((((.((....((((......))))((((..((((((........(((((((((((.............))))))........))))).(((((..................)))))...((((((......)))))).(((((((.....))))))).......(((((
(((..((((((((....(((((..((((.(((....((..((((((...(((((((((....(((((((....(((.(((((.......))))).))).(((((.....((((((.............))))))....)))))......................(((....))))))))))....))..)))))))...
))))))..))..................((((..(((......)))..))))....((..(((((((....(((((((((((.(((.......(((((((........(((((....((((.(((.((((.....))))))))))).)))))....(((((((((.(((((((((....(((((......))))).....
))))))))).....(((((((((..(((.(((....((((((......(((((..........(((((...))))))))))))))))....)))))).)))).)))))....))))).))))....)))))))......)))...))))........((((((.....)))))).......(((.(((((.....(((((
(.(((((((.......................)))))))..)).))))...))))))))........))))))).....)))))))..)).....................(((((.....))))).((((((..............)))))).......(((((....)))))........((((..(((((......(
(((...))))......))))).))))(((((((((.........)))))))))...((((((......))))))..((((..((((......))))...))))......))))))).))))).)))).))))....))))))))...........))))))..))))................)))))))....))))))
)))))..((((((((((((((((.((((...........(((((((((((.((((..(((..((((.((((((.....)).))))))))..))).)))).)))))))....))))))))..............((((((...........((((((((.((..((((...))))..)).)))))))).....))))))))
)))))).))..)))))).(((((.((...)).)))))))))))))))..))))))).)))).................(((((.......))))).................)))))))))))))))))......))))).)))))..............(((((((((((..(((((((((....)))))).)))....
((((...))))...(((((.(((....))).))))).((((..((...))..))))...............))))))))))).............((((((....................))))))....((((.......)))).(((.........(((.((((......)))).))).........))).....))
))))))))))((((((.(((.((((((((((...................)))))))...(((((..............))))).)))))).)))))).......((((((....))))))
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-511.19] kcal/mol.
2. The frequency of mfe structure in ensemble 2.48614e-29.
3. The ensemble diversity 498.82.
4. You may look at the dot plot containing the base pair probabilities [below]

III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
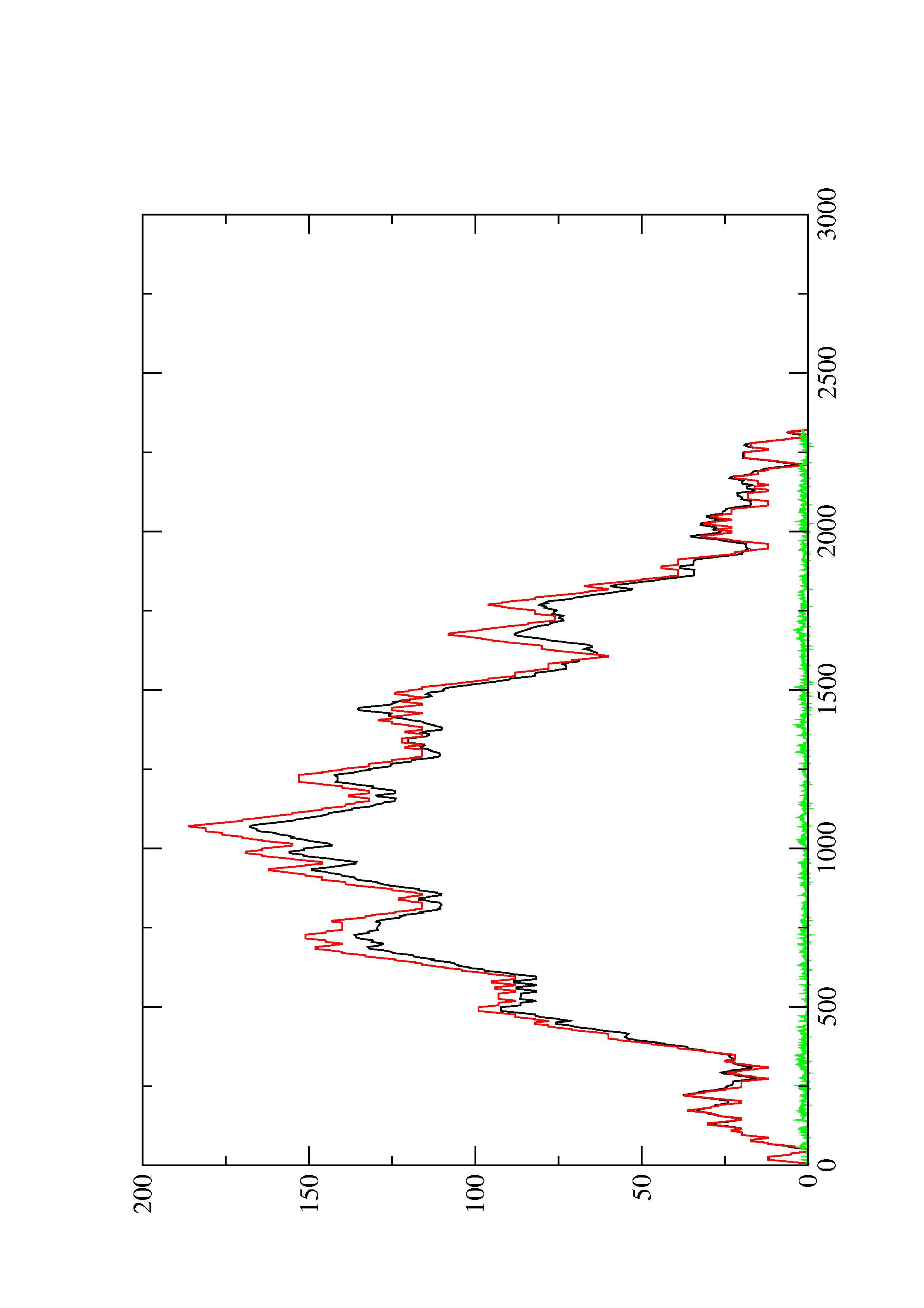
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.