lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01044148.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-602.59) is given below.
1. Sequence-
CGAAAAGUGCAAAAAGUUCAAUAAUAAGAUCUUAAAGGUCGAAUAAAUCAAAUUUAAAUA
AUUAAUCCAUAGCUGGUUAUGUACAUGGCCAAUAAAUUUAGUCUGUUAACAUAAUCAAAA
UUGCAAUUAGUAAAACAAAUUUCAACAUAAAUGACCAAUAAUGUCAAUAAUAAGACCAAA
CAUAGCCUUCAAAACUUUGGGAUGAUAAAUCUAAUUCCUCUAUCCUUCAGUACUUAAAUA
GCAGAUAGUAGUUUUCACGACGCAGGCCUAUGUCUCGAUGAGCUUAGGUCACAAGUCCAU
CAACAUCUUAGUUUGACUAUAAAUGAAAUUAAAGAAGGUAACAAAAAUGGUCGAAUCUUG
UGAAGUUAACUAAAGAGAUGUAUAAUAUACCAAAAUGUUUUUUUUUUUUUUUUUUGGAUC
UUGCAGCCCAAGAUGUACCAGGUGAGAUAUUGAGAUUAAAGAGGUUACUAAAUAUAGAAG
UGGCAUCUCUUUUAAGCAAACCAAAAAAGAAAAACAAUAUAUAGCCCCUUUUGUUCCAAU
UUACACGGCAAAUUUGAUUUUGUUCGUUCCAAAAAAGAAAAAUGCCACCUUUCUAUAAUU
AAAAACUUAAAUUUCAAAAAUCGCCUUGUGUUUUAAAUCCCAAAUUUAGGAGACUUCAUU
UUUUUUUCUUGAACUUCAUGAAAAAUUGAAACGCACAGACGAUGGCGGAGCCAAAAAUUU
GAUAAAGGGUGUGUAAAUUUUUUUUUUUUUUUUUUUUACUUAUGUUAAAGGUAUUCAAAA
UUAAAUACUUGUAACAACUAAUAUUUAACCUCUAUACUCCACAUAAUAUUCUGACGAAAG
GUGUGUAUACUUAACCACCCUUCGACACUGCACAAGGAGGGAGUUAACACAAGUAUCAUG
UAGUUAUAUUCGUACUGAUUUAACACUAGGAGAUUCUUCUCAUCUGUUCUAGCCUUGGUG
AACAGAAUGACCUGGUACCUGUUGCUGGUGGGAGAUGACAGGUAUCCCGUGGAUUUAGUC
AAGGUGCGCGAAAGCUGGCCCGAACAACCACGAUCAUAAAAAAAAAUAUUUGUACUGAUU
UUGUUCAGUGACUAAUGCAAUGAGAAAACUAUUUCCUUUAUUUUUUAACUAUUAUUAUUA
UUUUCUUCUUUUAGUUUAAGAAUCUAUCUCUACAUGAUUGGUGAGAAAAAGGAUUCUUGC
UUUGAUUCGUCUGAGAAAUGUUGCUAGAAAAUGUCUCAAGAAUUCCUGGUAUUUUCGCAA
AGUGAAAUCGUGUUAACUUGGACAUACAGUCUGAUAAAAGUAGGACGCGUAUUUCAUUUU
GAAUCGGUAUUUUUUGCAGGAUAGAGGUGUCAUAUAUGGCCAUUAGCUGUGUCCUACGUA
UAUAGCCGGACUUCCACCCCUUCUCCAUCCAAUCCCUUAACCCCAAACAUUAGUCCAAAU
UACCAUCAACGGAGAGGUGGGCUACCCCGUUGCAUCGAUGCAAACAGUAGUCCAAAUAAG
AAAGCAGUCGGAGGGAGGAUAAAAGACGCGUACCAGAAAGGCGUAACGCGACCCACACCA
UCAUCAGCAUGUUGCAAUACCUGAUUGUUCCUGCGUCAAUAAUAACUAAGAUUGGUAUCA
CGUCCUGUCUAAUGAAAGAACAACGAAACAAAUAUAUGUUCUAAGAACUAAUUUUGUCAU
ACUAAUUAGAUUUAUCAACUUGAAUCUACACGUGACUGCCAAGUUUCUAGCUUUAAAACA
UUGUGUAAAUUUGACAAUUCUAGCCAUAUGUGAGUUCUACCUUUUGCCUAGAGAAACCUG
UGAAGGCAUAUUAGCAAUUUGAGCAUAUACAUGGCUAAUGUAGCUCGGAUUUUUCAAAAU
UAUUGAUGGAUUGGAUUGGAUUGGAUCCUUCAAAAGUAGUGUCUUUCUACAGAACCUGAA
AUAGGUAUAACAACAUUUUUGGAGAGUCCGAACAACAUGAUACCACCCGCAUUCAUUGCU
AAAUCACUCUUACACAACGAUGGAACAUCUUUGGAUGUUAAAUCAGGGAAAACUCUCCUA
ACAUGUUAGACAUCAUAUGAUAGCAUUUGGUAUCUAUGUAUAUAAUAUCGAGACAAUAAU
UCACAAAUAACACAGCAUAUCAACAAGCUGACGUAAAACUUGAAAAAAAAAAAAUAGAGC
UAUCACAUAAUGUUGAAAGAAUACAUAAUUGCAUUCACAACACACACAUGGACAAGAUAU
CUCAACAUAACAAUCAAAUAAACUAAAAAGUACUUUGAAUUCCCGAAAUUGAAUGUGGUA
UUUAUUUUACUUGAAAUGUUAAGGUAUACUAUAAUGUAUGUAUGUCUGUACACGAUAUAU
ACUUCCGAAAGAGUGCCUUGAAAUUGCAUAAUAAAGAACUACCAGUUUUCGAAUUCUUAA
AAGGAGAAACUCAUCUGCAACUGUCAUAUCUACAAGUAUAUAUAUGCAAGGGGCACACAC
UCAUUGUCCUUUUUACAAGAAUGCGUAAACCCGGAAGUACGUGUAUGCAUCUGGUACCGC
CCAACUAAGAGGUGCAAGCGCAAACCCUGAAAUGGCAUCAAAGUGGCAAAACGAAGUUAA
UGACAGCACUGUCCUUAAUACACUAAAAUUUGCAAUCCUCACAUUCAUUUUCAAAUUAGC
AUUGGUUCUUCCUAUGUCUUCUACAUGCAUUGUGUAACGUUAUCGGGAAUUCGUACAAAC
CCAUCUACACACACUCUCAAAGUCGCUCAGGCAGAUCAUUCCUUUACUAUUGCAAA
2. MFE structure-
.......(((((..(((..........(((((....(((((........((((((.....((((((.((....((((((((((((.(((.((.....)).))))))..)))))))))....))..)))))).....)))))).........)))))......(((........)))........((((.....(((((((
((((...(((((.......(((((...................)))))(((((.....((((((((..((((((((....((.((((.(((((((...............((((((((((...((..(((......)))..))...)))))))))))))))))..))))))...)))))))).......((((((.....
...........))))))((((((.....))))))(((..((((.((((((((.....((((((((((((.........))))))..))))))((((.(((..((((((((((((.(((.((((..((((((...((.(((((....((((..(((..(((((.......)))))..)))...))))..............
...................((((.((((((((((((.(((........)))...........((((((.(((...))).)))))))))))))))).)).))))(((...)))..))))).)).))))))..)))).))).))))))))))))..))).))))(((((.(((((((........))))))).)))))..))
)))))).))))...))).......(((((.((((.((..((((((.....(((.((.(((((((((..(((....((.(((((((..(((....(((((((((..(((...................(((((((..(((((((.(((..(((..(((.((((((((((.....(((((((((((((......)).)))))
)))))).(((((......((..(((..((....))..))).))....)))))...(((((........)))))....)))))))))).))))))..))))))))))....)))))))..((((((.(((((....................))))).)))))).)))..)))))))))..))).(((((((((((((((.
...(((...(((.((........)))))....))).)))))).))))....))))).(((((((((((((((((.((((((((.....)))).....))))..)))))).)))))))))))...((((......((.((((((.((..((((....))))......)).)))))).))......))))))))))).)).(
((((((............................................)))))))((((((((....(((((....)))))...)))))))).........)))))))))))).)).)))....(((((.((.....))....)))))...)))))).)).)))).)))))(((((.....))))).))))))))...
...))))))))))((((((..((((((((((((...((((((..............))))))..(((((......(((((......((((((((......))))))))...))))).....)))))..............((((((((...(((......(((((((.((((.((((......((((.(((.....(((.
...)))....)))))))...)))).)))).)))))))((((...(((((((((((((((...((((.(((((.(((...))).))))).))))..((((.......))))...(((((...)))))..........)))))))))))))))...))))..........(((.....)))....)))...)))))))).((
(..((((((....))))))..))).((((......)))).....)))))))).....(((((((..(((((((.(((....))))))))))........((((......((.((((..........))))..))......))))..............))))))).....(((((...((((.((....)).)))).)))
)).......))))..))))))............................((((...((((((((((((.(((((((((....((((((....))))))(((((((((.((.....((((((((((......)).)))))))).......)).))))))))).((((((.........(((.((.((((.((.((((((((
(((((.((.......(((..((..(((((.((......)).)))))..))..)))........)).)))))))...)))))).)).))))..)).)))..((((.((((((((((........)))...))))))).))))...........((.(((....))).)).......(((((.((((....)))).))))).
..........))))))..))))))))).))))......((((.((.....)).))))....(((((.....)))))........))))))))))))...............)).))))))))).....)))))))))........))).)))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-649.82] kcal/mol.
2. The frequency of mfe structure in ensemble 5.22324e-34.
3. The ensemble diversity 476.55.
4. You may look at the dot plot containing the base pair probabilities [below]
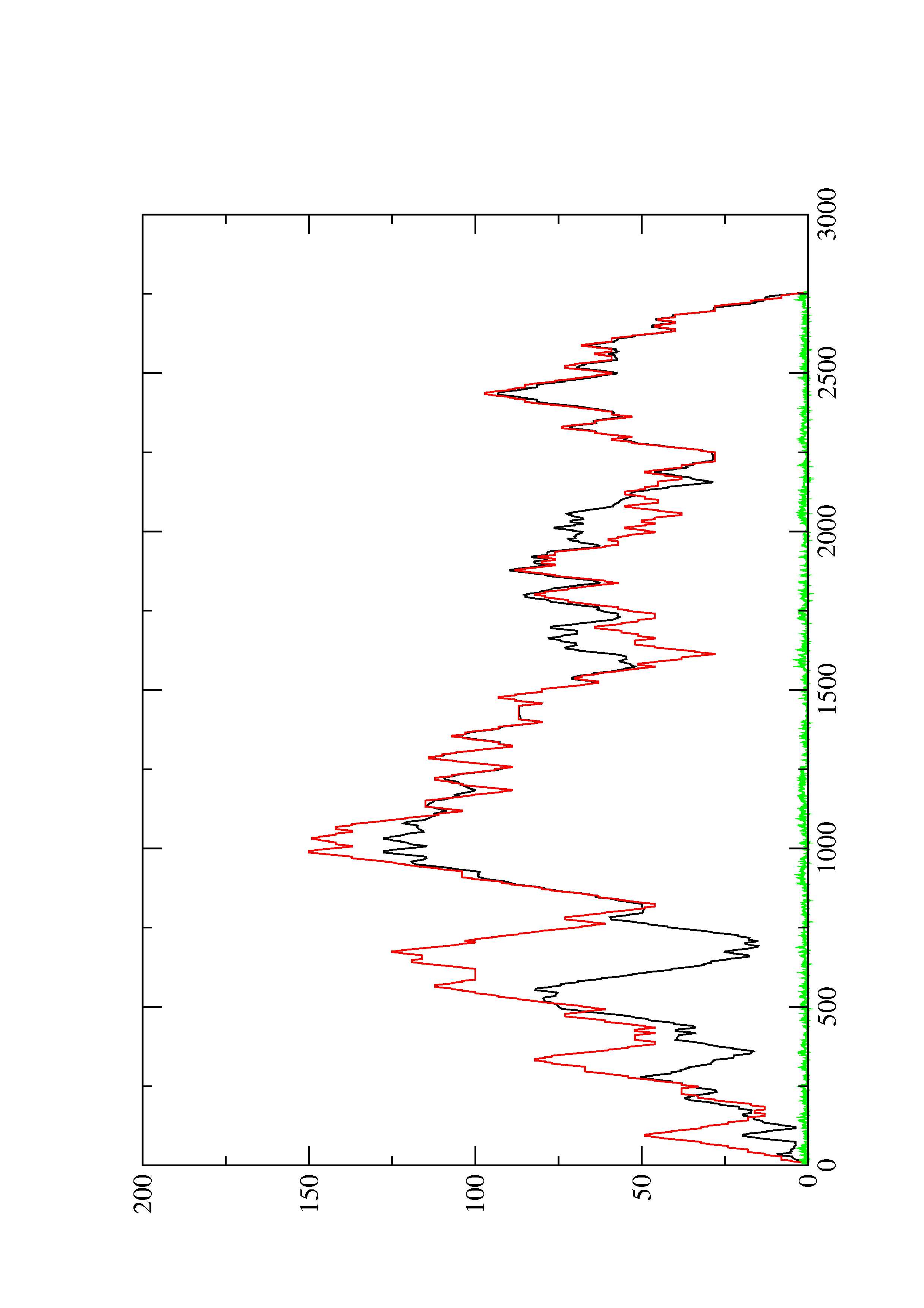
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.