lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01044440.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-712.50) is given below.
1. Sequence-
UUCAAGAAGGGGGAAAGGAAGGAGAAGAGGAGCGCGAUUCGAUGAUGAUAAUUGGGCGUC
UGUGGUAGAUAGAGAAGCAGAGGAGAGGUUAUUGAAGGGGAAUGAUGAGGAGUUCAAGAA
GGGGGAAAGGAAGGAGAAGAGGAGAGGUUAUUUUAGUGACGAGAGCGAUGAGGAAGAUUG
AUUGUUUAUUUUUUUUGUGGAGUUUGAUGAGGAAAAAUGGGAGCUUUUACGCUUGUUGCU
GAAGUUGAGGAAUGAUUUGCAAUGGUGUUAGAUAGGUUAGUGAUGAGGAAGAUUGAUUGU
UUCGCUUUUGGUGGAGUUAGAUGAGGAAAAUUGGGAGCUUUUACCCAUGUUUCUGAAGUU
GAGGAAUGAUUUUAAAUGGUGUUAGAUAGAUUAGUGAUGAGGAGAAGAGGAAAUGUUGUU
UUAGUGACGAGAGUGGUAAGGAGGAUUGAUUGUUUAGUUUUGGUGGAAUUAAAUGAGGGA
AAAUGGGAGCUUUUAAGCUUGUUUUUGAAGUUGAGGAAUGAUUUUCAAUGGAGUUAGAGA
GGUUAUUGAAGGGAGAUAAUGAGGAAUUCAAGGCGGGAGAAGAGGAAAGGUUAUUUUAGU
GAUGAGGAAGAUUGAUUGUGCGGGGAAAAUGAAAGCUUUUAAGCCUGUUUCCAAAGUUGA
UGAAUGAUUUACAAUGGUGGUAAUUGAAGUUGUAAGUAGAUCUGUUUGGGAGGUCUUGCA
CUCAACACUCCUGAACCUGAAAUUGCAAGAAUUGCGUCUUUUUGCUAUUGUGCAGUCCUU
AAUAUGUCUUAUUGAAUCCAUCCAGUGGCCUUCUCCAGGAGAGGCAUGAAAAUUGAAGCA
UUUGAGAAUUCUGAGUAGUGGUCUUCUCCGAAUACACGUGAAAAUUGAAACAUUGAGAAU
UCCUAAGUCGAGUAAGUAUGGGGGUGUAUUUGAUUAUCCAUUAUUUUUUAUCUGUGCUUC
UCUUGGUAACCGUUGUUGACACUGAAACGCUGGGGUUGUGCCACUAUUGGCGAGGUUAGG
AACCUUGGUAUGACACGAUAACCGAAAAGGUGGCCACAUUCCCAAGGACAACUCACUCAC
UUCUUAAGAGGAAAUACUGUAUUAAAAGAAGCAAAUAAAAAGCAACGCUUUACAUAAAAA
UUUUAUACUCCCUCUGUUUCAGUUUUUGUAACUGGUUUUAACUUGACACAACAUUUAAGC
AAGCAUAGAAGUAUUUUGAAUCUUGUGGUCUUCAACUAACUAUUCAGUACAACAUCUUGA
UUUGAGCCAUAAGCUUCUUAUAUAUAGCUUAGUGCUUUAUUUGCGUCUUUUAAGAGAAUG
CAUUUGCUCUUAAAAAGUGGGUGACUUAUCCUUGGAAAUAGUCAACCAAGGUGGCCUACU
UUUCGAUUAUCGUGUCACAGUGGGGUUCUUGCCCUUGCUAAUAUUGGCAUAACCCCGACA
UUCAGUUGACCUCAGUUUUUAUUAGUUAGUGAGAGAGUGAAAGAUAAAGGCAGAAAAGGA
ACCACCAUCAACAAUAGCAACUACGCCUAAAUCUCUAAGUAGUCGUUUUUGGCUAUAUGA
AUCCUCUAUAUUCAUUCAAACCCAACAUCAGAUAGGUCACAUGAGUAUCUUGAUAACUUU
GCAGAAUAGCUGUUUUUUCUGCAGCUUGGUAAUGUUGUAACUAUAAUCAGUGGAAUUUCA
UUUCUGUAUACCCACUUAAUACUCUAAAUUGACAUAUUCAAAUCAACUUACAUUUUUACC
AUUUUCACAUGAUUACAAUCCAGCAAGGCGACUAGUUAGUAAAUAACCAAGAAUUAAGGC
UGUUUUCUUCAACUUGCACUUUCUUCUAGACUUCAAAAUGUGCCAGUGACACCAAUCUUA
GAAGAAUAAGUUAUGAAGAUGCAAUCUCUCUUGCAUUUAUAACUAGGUGUAAUUUUAAUU
AAACUGCUCCAAUUCUGCAGCAGUGUGCUUAUUAUCUUGAUGUCUUAAUUUCAAUGCUCU
UUUCAGAGUUUGCCCCCAUCCUCUGCCAAUAUGUGAACUUGACAUGCUUCAUUUUGCAAU
CACUCUUGGUGCCCUGUUAUACAUAGUUCCUUGCCAUUCAGCUCAUUGAAUAAUGGGCAU
CUAUUCAGUACUUAUAUUAGCAUCUAUAUCUGGAGGCUAGUGGCUUUUCUUAUCGACUUG
UUUCUCUUUUUGAUAGUACAGGAGAGGGUUAUGUGGAAAGUUUUAGCUUUUGUCGAGUAG
UAGUACAUUGUGGUUUGGUGGUGAAGAAUUUGAAUCAUCUGGUUUUGGAGGCAGAACUUC
UGUUCACGAGGUUGCACCAUGUUUGUUGUCAUUGUUAUCUGCCCUCUGUUCUUCAUCAUU
UUGGGUAAGGGAAUAGGAGCUGGUUGGAAGAUUUUAGGAUUGAUGUGUUAAAUUUCAGAA
UUGCUUCAAAUUUAUCUUCAUUGGAAUAGAACUGUUACUCCGCACCAUAUAUAGGUACCC
CAAGCAUGUCAAUACAAUCCAUGACAUGCCACUCCUGUUUCUUGGUAGAUUAACAGCUUG
GCUAGUUAGGUAGGUAGCCCUUUAGUUGAGGAUUUCUGUACAGUGUUGCUUGUCAUAUCA
AAACUUCAUGGUAAUUUAAUGUGCACAGAAAGGAGUAGAGAUGAUGUUAUGCAGAGGGAA
AACAGAUGAUACUGAUUUAUUUAAUUGAAACAAGAAAGUUCAUGAAAUUUGCAUUCGUGG
G
2. MFE structure-
.(((((((((((((.(((..((..(((((((((((((...(((.((((((((.(((((.(((((((....(((((((((...(((....(((((.((((((.(((........)))....(((((.(((....((((((((..(((((((....((...(((((..((((((.(((((((..((((...........(((
((((((..(((((((.....((((......))))..(((..(((((((((((........((((((.((((...(((((............((((((((..((((((.((.((((((...(((..((((((((((........))))....(((((.((((..((((((........((..(((.((((.((((.(((((
(((((..(((....((((((....((..((..(((((.(((..((((((.((((..(((((((((......(((((.((.(((...(((((....)))))...)))...((((((((....)))))))).......((((((((((((...(((..((((((((.....((((..............(((((.....(((
(.((((.((((((......(((((...................)))))(((((((...(((.....(((((((((.............)))))))))..)))..)))))))))))))...)))).))))....)))))...((((((..(((.(((......))).))).))))))))))......))))))))..))).
..)))))))).))))((((((((((((............(((((.(((((((.(((.((..((...((((......)).)).....)).))))).))))))).))))).((..(((.(((((((((........))))).)))).)))..)).))))))))))))....)).)))))..)))))))))...(((((((((
(((.(((((((((((.(((((((((((...(((((((((((.(((((((..(((((((((.(((((((.((.((((((((((.(((((((.((((......((((((((.((((((((..((((..(((.((.((((.......(((((...(((((..(((((.....)))))((((...(((((........))))).
))))))))))))))...(((.((((((((..((((......(((.((........)).))))))))))))))).))))))).)).)))....)))))))))))).))))))))........)))).))))))).)))))))))).)).))))))).)))...........))))))..))))))).)))))))))))...
))))))))))).))))))))))).)))))))))))))))).))))))..)))..(((((((((..(((.........)))..))))))))).........)))))..))..))..))))))....)))...)))))......))))).)))))))).)))..)).......)))))).....)))).)))))...((((.
..((((((....(((((.((((.....((((((.......))))))..)))).)))))..........(((((.....((....)).....)))))..)))))).....))))...........................)))))).)))....))..)))).)))))))).))))))))...))))).....)))).))
)))))))))))))))))).))))))))))))))))............)))).))))))).........((((((((..((((((......))))))))))))))..................((((((.((....)).))))))...)))))).)))))....)))))))))....))))))))....))).))))).))
))))..)))))......)))....)))))).)))....)))))....))))))).))))).))).)))..))))......(((((((....)))))))................(((((((.((((....)))).)))))))))))))))).))..))).)))))))))))))((.(((..((((((((((.........
...)))))))).))..))).))((((.((((((...((((((...........)))))).(((((((....))))))).....)))))).)).)).(((((..(((.((((.(((((((((((((((((....(((((((((((((((.((....(((((...((((((...(((((.((....)).)))))))))))..
..)))))...(((((((.....)).))))).)).))))))...(((.......))).......((((((((..........)))))))).((((((..((((..(((.((((((((((.(((((.((....)).)))))....)))))..(((....(((......)))..))).(((((........)))))..)))))
.)))..)))))))))).)))))))))....))))))))....))))))))).)))).........(((.((......)))))........)))..))))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-763.74] kcal/mol.
2. The frequency of mfe structure in ensemble 7.79265e-37.
3. The ensemble diversity 669.52.
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
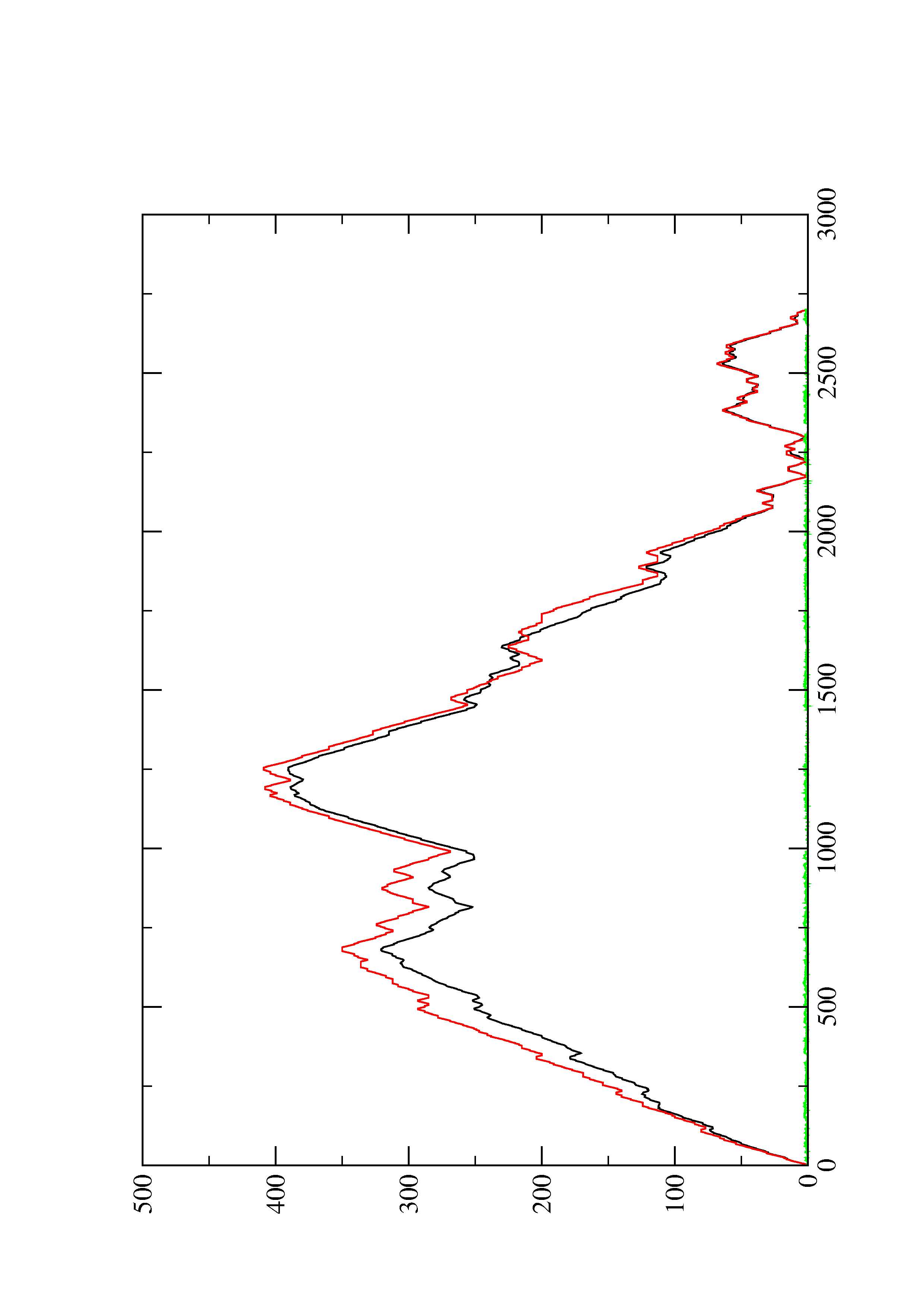
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.