lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01046549.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-522.17) is given below.
1. Sequence-
CUUUUUAUAUUUUUAUAUUUUUUAUAUAUAAUUAUUAUUAACAACUCUCGUAUAUUCAUA
CUAAUUUUAAAUUUAAAAUAUAUAAAUAUACAUUUUGAAUUAAUAAACGCGUUUAAUUUU
UUUUUUUUUAUAAAACGGGUUGGCUCGGACACUGUUUACGUGGGUUGGGCCGGUCCGGAU
CAACCAUUUUGGACCGGUUUGGACCGGCCCAUUUCAACCCGGCCCGGAUUGGGGCAGCCC
GGACCUGACCGGAGCCCGGCCGGGCCAUGGACCAACUGGUCCGGACUGACCCGGACCGGC
CCGGAUUACAGGCAUAACUUCAACUAACAAAGAGACAGAUCCUAACUGGACCCUUAAGCU
AUUUAAGGAAAGCAUUUUUAUAUCUAUACACUCACACUACCAUAAAAAGGAAACAAGAAA
CAGCAAUAAGAAUGGCCUUUGUUUUAGCUCUAAACAGUGAUGAAAUAGGCUGGCUCAAUC
ACACAUUACUCAUUGAGACAUACAGGAAAUCUAACAGGCAUAUAUAAUAAAACAAUUAGU
AUAGCCAUCCAGUUCAUGACUCAUUACUAACAGGUUGCCAUUUAAACAUCAGGCAAUCUC
AGCAUAUGUCUAUGCGUUCUAUUAUCAGUUCCCAGAGAAUGACUAUUAAAUAGGUGAAUG
CAGCUUGCUUAAGAAUAAUAGAUAAGAGAGAAAACAUAACAACUAUCAACUAGCUCUAUU
UUUGCUUAUGCAUAUUCAGAGUUUCGAACAGCAAAUACCAGCAGUUGGCACCAUACCACU
UUAAAAAUUGUAACUGUAGGAAAAUAGGAUGAUGGGAGUAGCCAGCACCUAGACAGGUUG
UUAUUCCUAAUUUUCAACUCAACAUGCUUGUUUAUAUCUUCUCAUGGUAGAGAAGGGAAA
GAAACAGUCCUUAAAACAUCAAACUAGUUACUAUGCAUCGACAUGUCCUCAAAAGAAUUG
CAGAUUUGAUUAAAGCAUACAGUGAUUGAUUCAACUAGACUGCAAGCACCUAAGGAGAGU
GAUUUAUAAAUUUAAAGCAUGCUGGUCAAUGGACGCUCUAAUUUCAACAUCAGACAACUU
CUUUGAGACCUUUACAGAGAUCAUGCAGGUAACGUUAACAGGUUACCAUCAUUAACAGUU
CACAAAUAAAUGUGACUGGCAAGUUCAAGUUCUCAAAAAACAGAUUUGGGCCUGCCUUAC
UACUCAAGAGAGGGAGAGCUCCAUCAUUUAGGAACAAAAUAGCAUGAUUGAAGCAAGAGA
AUCUUAAACAGGCCACUGUACUAACUUACCAUCAAAAUUUAGAUUUUAAUCAAAGUCUAU
AUUACCAUGUGAGCAAGGAAAACGAACUGAUUUGUUGAGAGAAAAAUUUACUGCCAAGAG
AUCAUCAAAGAUGGGAUCUUUACCCUAUUUCAGUUUAUUACAUGCUAUGCUCAUCCAACA
CAUCAGAAAAUCAGGAGAAAAACAAACGCCUCCCCCAUUUGACACACAUGUGCACACAUC
UCAACUGACCGACAAAGCUAGACAAAAGUACGAAAACUCCUGAUUUCCACACUAGGUAUU
CAUUUUCAGGUUGAGCAUGUAUGCCUACAGCAAGCAACAGAACCAAAUAAGGGUGCAUAA
UCUCCGGCCACACUUUCACUCAACCAUUCUCACUCUAUUCUAUUCAUAUAGGCCUAUGGU
CCCAAUAACAUAAACAAUAAAAAACAGCAGUUGUAAUGCAUUAAGCCUCAGCCUGAAGUA
GAUGCAAUUGCACUUUAGAUUAGCCAUCCAAAGCUGAUAUUUUCCAGCCCCUUUUCCCUU
UAAAACACUGAGCUAAACAUCAUAUUGACCCAAAAAGACUUCACAUUUAAGGCAUAGAAU
GGCAUAACUCCAACCAGCAAGUCUUAAACACACAUCUAUCCUAAGUUUAAGUUCUAAAUU
CAGAUUUUAACCAAGUCUAUAUUCCUACAUAAAAGAAAAGAGUACUCAUGAUCCAAAACU
CUUAAGUACUUGUUGAUAUAGGUAAAAAUAAAGGAACCAAUGUAGGAGCUUUAUCAUGCU
GAAAUUGGAAACAUGCAGAGGACACAUAGAUCUACAGAUUUACUUCAAAGACUACAACUU
UAUCACAUAGAGAAUAGGGACUGCACCAAAUAACUUAAAAAUUCUAUAGGAAUUGAUUUU
AUAACAUGAAACGCGUAUUUCUAACCAAUCCAAAUCAAACAGAAACAACACUUUUUUCUU
AAAGACUCGUACUUUACAUGUCACUACACUGAUUCUUAUGUUAGAAAACCUAUUUUUCAC
UUAAAAAUCGCCUACUGGACUUAUCAAGGUUCUUAAACAUGCUGAUACCUAAGAUUUAAU
ACCUAAAUCAUCACAAAAUAGAUACAUAGACUGGAUGCAUACAGUAUUGGCAACCUAUAA
GCACAAAAUCAUGGUAACAGACGCACAAAACCAGCAAUCAUAUCAAUAUCUCACACGAAA
ACAGUAAGAAAUAACAAGAAA
2. MFE structure-
(((.((((.(((((((..(((((...........((((.((..(((((((((((((.(((...((((((....)))))).))).)))))))....(((..((((((((((((..................(((..((((((((.((((.....((((....((((((((((((((((((((..(((...)))...)))))
))))))))))).....))))(((((((....((((...((((......)))).))))..)))))))..(((((....)))))))))....)))).))))))))..))).(((((((.........(((((((.......(((.....))).((((((.....))))))....(((((((((..((......((......(
(.......))......))...))..)))))))))..)))))))............((((((((.((.((((((((((((............))))))...................(((.((((((((......))).))))))))..)))))).))...))))))))...((((((((............)))))))).
....)))))))))))))).))))))))((((.....))))..........((((((..((((((((.((((...........)))).))).....................((((((.....(((....))).....))))))..((((((((....((.(((((((.((..................))))))))).))
......(((...((((((((((....((((....)))).))))))))))...))).........))).)))))....((((((......))))))(((.........)))..............(((((((((((((((((...(((..(((.....)))..))))))))....))))..))))))))...........)
))))..))))))..))))))..)).))))..........(((((((...((..((.(((..((((((((((((.....((((((((((.((((.....((((((((.((((((........))))))........((((.((((......))))))))....((((((((.((........))))))))))((.((((.(
(.....)).)))).))...(((........)))........))))))))......))))..)))...((((....)))).....(((((......(((.((((((((....)))))))).))).....))))))))))))......))))..))))))))...........((((((((((((.(((....)))))))))
)...........((((((....(((((((((((((((..........((((((((((......................((.(((....)))))....((......))........(((.......))).......))))))))))......((((((((.(((((.((((..(((((((.(((.....)))......((
((((..((((((((((.........((((.........................................))))...((((((.......................(((((((((.(((.(((.((....)).))).)))..))))))))).......((((((........))))))............(((((....(
(((.....(((........)))..))))....)))))......(((((........)))))((((...(((((.(((((((.(((((((...............)))))))..))......((((((.....))))))...(((((((((.......((((((..............))))))...(((((((....)))
))))...............)))))))))........)))))...)))))...)))).((((.....((((((....))))))))))...........((((((...))))))....)))))))))))...............((((((((...(((((((.(((..(((((......((((((((.......(((((...
(((((.........)))))....(((..(((...)))..))).......))))).....)))))))).......))))).)))))))))))))).)))))))))...))))))...)))))))...)))))))))..))))))))....(((........)))........)))))).....)))).)))))....))))
)).........)))))..)))))..))..)))))))....................)))))..))))))))))).)))...
You can download the minimum free energy (MFE) structure in
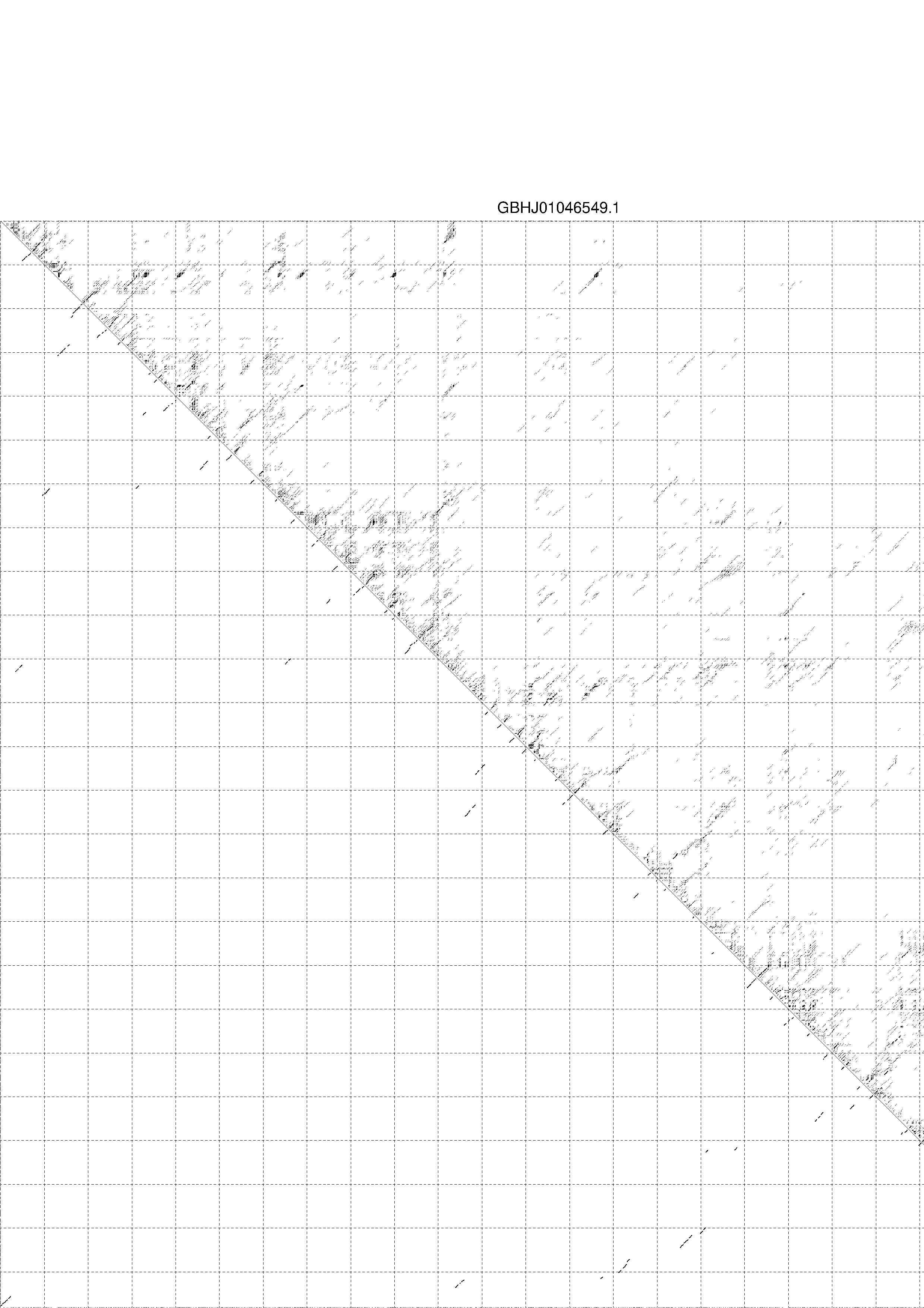
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-567.27] kcal/mol.
2. The frequency of mfe structure in ensemble 1.65678e-32.
3. The ensemble diversity 555.13.
4. You may look at the dot plot containing the base pair probabilities [below]
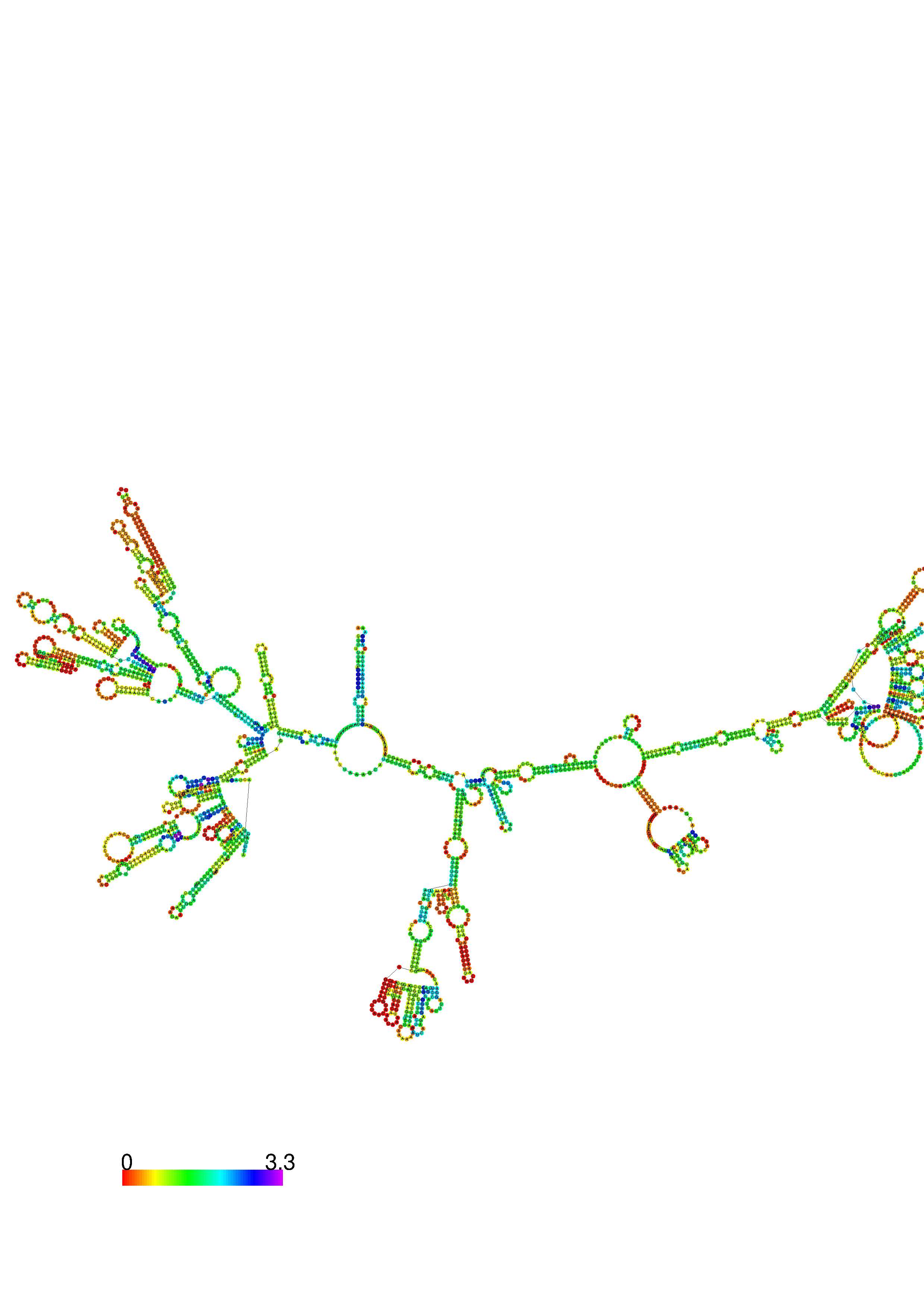
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
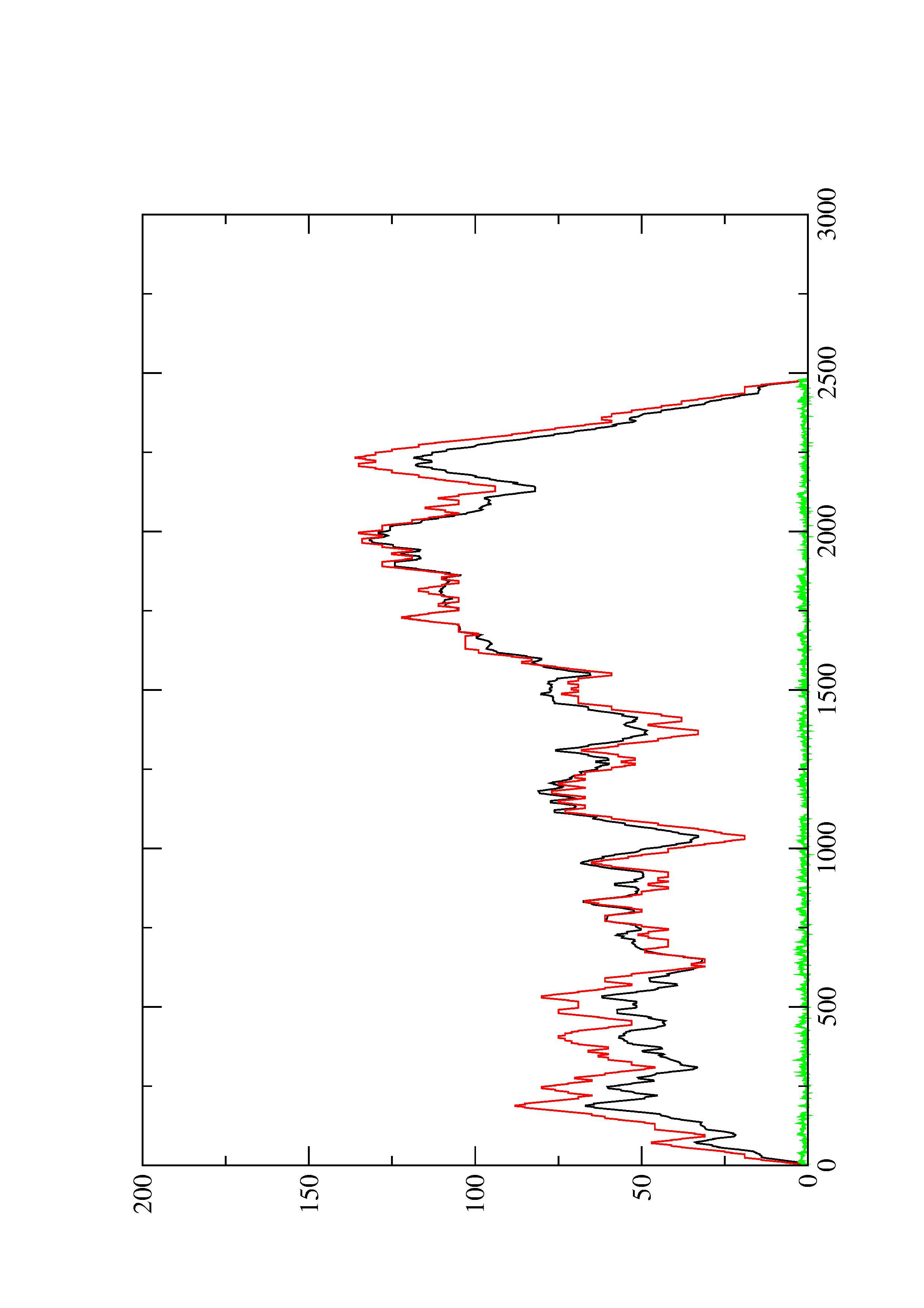
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.