lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01046705.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-777.90) is given below.
1. Sequence-
UUUUUUUUAAUCGAAAAAUCCCACGUGAGCUCAUGGAAUCCAACAUAAUAACCCCACGUC
CGGAGAGUUUCUCAAUACCUUAUCAAGGUAAAGAGGAUACAUGAAAAUGUAACUGUGAUU
GAUUAAAUAGCAUUUUGGGCUCAUGUCCUGCAUUGGCAAACGUAGUACUGGGAUGAGGUG
UCAACGCCGAAGCUAGAAACGAAAAAUAAUUCUCAUCCAUUGAUAUUAAAUAAUUCUCCG
GAAAUAAUAUAUAUUCAUAUGCUCAUAACCUGCAAAAAAAAUAAUAUAUAUUACAUCAUA
UAUCAAUUUUAUAAAAAUGUCAUAAUCUUAUAAAUGUCCAUGCUUCUUUGCAAGUUUGUA
AGUUUUUCAUAUCAAAGUUUCUACUACUUGUCAUAAGAUAGAGACUUGAAAACCUUUCUG
UGAACAUUAACGAUUUUCACAAGAGUCAUGAUUUAGUGUAGAAACAUUAAAAUGAUGGAA
CAUGUAGCAUGGUUAUGAAUUUUGUUCAAGACAUGAGGAUUUUCAUAAUUUUAUAUAUUA
CACCAUAAGAGUGAAGAGCAAAACAAAAUAUCACAAAUCUUAAACUCAAAUCAUAAAAUC
CUUGUUUAAAUUAUAAUAUAUAAUCCCAUGAACUCCUGCAUACAUUAGUUUAAUGAAGUC
CUUGAAGGAGGAAAGGUUGAGAAGAACAACCUUGAACCCUAGAUGAUAUGAAUAUGGUUA
AUGGAAGCAAACAUAAAAUAUGAGAAAACAAAGGAAUACAAAAAGAUUGAUAAAAUGGAU
AAAUAGAUAGACAUAACUAAGGAUUAAAAGGCAACCACGAGAUAUAUUAAAGCUCAAAAC
CUCCUAAUGAUAUACCUACAAGCACCUAAUCUCUUACGUCGAUCUCAAGAGAAUCAAUGU
AUCAAGCAACCAACGUUUCUCACAACGACCAAGCACAAGCUUGAACAAGUCUCAUUCUAA
AAGAAAAACUCUCAAAUUGACUCUCACAAGAGUUCAACACAAACAUACUAUCAAAGUCUA
CCUAUUGGAUGAAAGGCACGAUGUUUAUACUAGUAUAAGUUUUACUUAUCAUACUUAGAC
CCAAAAGUAACCCAAAAUAUCAAAGUGACCCCUAAAGUUUGGGUCCUUGCCCAAUUAAAC
UCCAAAGUCUUCAAGCAUCUCUCAAGAGAUCCUCUAUACGAAGGCUUGGUCUCUAAGGUG
CUCAAAAAGGCCCUACAAAGGGCUUCCUUUCCACUCCAAGCUAGGCAUUUCUACGAGCUC
CCUUUCCUUAAGGAUUGUGCUGGUCAAUAUCGUAUCAUUCUCUCCUCCUAGUAAAGGAUU
UGUCCUCGAGUUCAUGCUUUGGAAAAACAAUAAGAGAGAAAUAAGAAAUCAUAUUCUCAU
CAUGGUAUGAGGGUUAGCAUAAGUAAUAUCAUGCAAUAGAGACAUCCUCAAAACAUGACC
AAGGAUUCCAAAUCAAGGUGAGACCCCGUAGUUUGCUUGAACAGAAAGAUGACCUAAGGC
CAUAGCUAAGUUAUCGUUAUCGGACACGUGAGAUAUCCCGGCGUUAGAGCAUUGAUUUCU
CCUUUCGUUCUUCUGUGCGAAGGCAAAGAAAGAAGAAAAAAGACAUUCAUUUCAAAACUA
UGAGGUGAGAAGAAAACAGAAAAGAAAGAGUUUUGAAGAAAGCUACGGGAAAGUCAAAUC
AUUUCUUUUUUAUCGACUAUAUUCAUAAAAAAAGCACAAGAAACUUAGUCUAACCUCAAG
CAAUUUUCACAUUUUAUACCAACAAGCUUAUCAUCACAGGUAAAAAGAUAGUACAGAAAG
UGAAAAAGAUCAUUAUGCUCAAUCAAGUUCUUUUCAUUUUUCAAGACAACCUUAUCAUUA
AAGCAUUAUCAAACACUUCUUUGGAAAUCAAAGCACAUGAGAACAACAUAAAAUCUCUCA
AGAACAUGUCAUCCUUUCUCUCAAUGAGCCGUGGGCACAUAAGAUGAAUAUUCACCACAA
UCAUAAAUAUUAUCAUAAGCAAAAAGAUAAUAAAUAAAGAUAGGAGUCCAUCCUACCAAG
UGAACUUGCAUAUUCCAGGAGGGUGACAAGAUUGGAGCCCAUACAACUUAAGCUCAUUGG
GAACAAACUAAGCAACUUGGGAGAUAUCGACAAACAUAUGGUAGGGUUUGGGUUCACAGA
AGUAUAACAAAUUUUCUACCCUAAUGAAAAUCUCACCUGCCGCUAAGGAUCAUUAAAAAU
AGAAGGAGAUGGGUACGUACCUUGAGGACAAAGCAUUGGGUUUGUUUUUUGGUUCUCCAC
AAGUGCAUUGCAAACCUCUUCCAUCUUAGGCAUCACUUUGGACCAAUGGGUUUGGUCCAC
CUUCUUUUCAGCAGGAGUUUCUUCGGUAGGUGACCAUUUUAAUUAAAAUAAUGAGUUGUA
GGUAUCACCUUUCAUUAUCAUGGUCAACCCAUGUGAAUUCAUUUUCUUCAUCCCAUAGAG
AAUUGUUAUAGACAUAGUAGCCUUCAGAGAGUGCUACAUCAUUGUUUUGUUUCAUACCAC
UAGAAACCUCCAUCGGAGGAAUCACAUCUCCUUUAGGCAGCUUUUGGAGCAAUAGCUUAG
UGCUGUGUUAGUACAACUAAAGUGUCAAGGAGAAUGUGGUACUGUUGUUUAUUUUCCUUU
UAAACAACUACUACAUUAAAAGAAACUGAAAUGUUAAAGGCUGAGAGCAGAAUCAAGCCC
UUCAUAGAAUUCAAAGUCUUCCUGAUAUUGCCAAAAUGACCGUCUCGGCGGGAGAGAGUU
AUUCGAUACUUUUAUGUGGGUGAGAGAUAACAAAUAUUCAGAAAGUAGUCGAGUUGCACU
UAUACUAGCUUGAACAUUACCCUUGGAAUAAAAAAUAGAGAGUGAAAACAAUACCAGAUG
CUGAAACAUGUAUGUCAACAUAAUGGAUGGCAAGUGUUUCGUGCAAUAAUUAAGUUUUAC
GUAAGUGCACUUAGUCAUACAAGUAAUAUAUAAGAUUUUUAAAUUCAGAUUUCACAUAGA
UCCGAAAGAUUGAUCAUUUAAUUCAAAUUCGUGUUUCACUAUUGGAGAAACAAACAAUUG
UGGAGCUGUUGUUAUGCUAAGAAAACAAUAAAACUUAUAGUAAUCUGAAAAUAUGGAGUU
UAAUCCAAGGAACAAAAUGACUAGUGCCUUGGCUUUUACAAUCCGGUUGAGUUUUUAUUU
CGUCAUGCCUAUUAAUUUUUCUGAGUUACUAGUUGCAGAUUAAUAUCACUUAUUGAGUAU
UUUUUCUAACAACCCUAAAACUACUUGACUAGUUGUAAUAUAUAUGUGGAUGUCGAUCGA
UAGUGAAAUACCUGUGUGAUUUCAUUUAUAUCGACUAAAAGGAUAAAGUUUAUAUUUCUA
UUACAAUAGCGGAUGGUUCUCCUUCCCUAUAUGGUUUAGAAACAAGCCUUACUUCUUAUU
ACACCCUUAAAUUCCUUUUUUCCAAGUUCAAUUAUGAGCAACUGAUAGUGUUCAACUAUU
UUGUCAAAAAAUUAAAUAAUUAGAAUCAAGAAAGAUAAAUAACUCAAAUAUAGAAAACUU
AAAUAAAUAUGAAUGAUAAUCAAACCACAAG
2. MFE structure-
....((((((((((......((((((((((..(((((..........(((........(((((((((((((((..((((((...))))))..)))))(((((....)))))((((((.(((....(((.(((....((((....))))))))))..))).))))))....((((((((((....))))............
(((......)))))))))..............)))))))))).......))).)))))..)))))).................((((((((((((.(((.((((((((((.........((....(((((((.((((...((((((((((((....)))))....((((((((((((((((((...((((...)))).))
)))))))......((((.((((((.(((...))).))))))))))((((.(((((((((....))))).)))).))))..((((...))))(((.(((((...))))).)))(((((((..(((((....((((((((((..........((((................)))).....((((((...............
...))))))....))))))))))....))))).)))).))).(((((...)))))...(((((....)))))(((((((.......)))))))..........)))))))))..........))))))).))))....)))))))........)).......))))))))))..)))((((........(((.(((((((
(((((((.(((((...((((((((....((((((((..(((......((((((((.....((((.((..(((((.......((.(((((......(((((.((((((......((((......))))..(((((....)))))...........((((...)))).(((((((...((((((((....)))).))))...
.....(((((((......(((((....(((((.((((((((((((...((((((((((.............(((((........)))))..............((.((((........((((((....))))))..........((((((((.((.(((((....))))).))......)))))))))))).))((((.(
(((....(((((((....)))))))..........((......)).........))))..))))(((....))))))))))))).))))))))....(((((((.(((.((((..(((((((....))))))).))))..)))..........)))))))...((((......)))).....(((((((((((((((..(
((((....(((((.....(((.....)))....))))).....((((...))))..(((..(((((((((((((.(((((((.....((((((....(((....)))..)))))).....((....)).((((.(((...((......))...))).)))).((((.(((((((.(((....))).)).)))))))))..
......((((((((......)))))))).....................))))..))).))))))))))...)))..))).....((((((((.((......))))))))))..........)))))..))))))).................))))))))....)))).)))))..)))))....)))))))..(((((
((((((.((...................))))))))))))).............(((((...(((.(((((.......(((((.....)))))...................((((((((((....(((.........(((((((((((.((((....................)))).........(((((((......
.....)))))))........(((((.....))))).((((....))))............((((..((....))..))))...........))))))))))).........))).))))))))))..............((.(((.(((((((((...((((((((((((.(((((((.....(((((....((((((((
((..(((((((.........(((((((((((((..........((((.....(((.((.((((((.....((....)))))))).)).)))...)))).))))))).((......)).((((((((.....)))))))))))))).........))..)))))))))))))))..............((((((((....(
(((...))))))))))))(((((.....))))).(((......))))))))....)))))))))))))))......((((((((....))).)))))......((((((((((.........((..(((((...)))))..))....((((((..((((.((((..((((....)))).((((((...))))))....))
))))))))))))(((((((((..((((((((.........)))))))))))))))))....))))).)))))......((((((........)).))))))))...))))))))).))).))))))).)))...)))))((((....))))..)))))))(((((((((((((((((..((........))..))))...
)))))).))))))).............)))))))))))...))))).)).......)))))..............)).)))).......))))))))......((..((((((((((..((((.(((......))).)))).....)))))).))))..))......................((((((((......((.
..((((...((((.........)))).)))).(((((((.((((..........)))).)))))))(((((((..........)))))))...))......)))))))).....(((((..((.((((((.((..........)).)))))).))..))..))))))))))))))))))))))..))))).)))))))))
...)))))...))).........))))(((..(((((((.((((............)))).)))))))..))))))))))))))))))).......)))))..)))))....(((((((((((((((((((.......((((((((.((((...((((((...((((((.(((.((....)).))))))).))...))))
)).)))).))).)))))...................((((((...((((((((((..((((..((((((.....))))))))))...........)))))).))))...))))))...............................)))))))))).)))))...))))..
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-844.12] kcal/mol.
2. The frequency of mfe structure in ensemble 2.16323e-47.
3. The ensemble diversity 764.06.
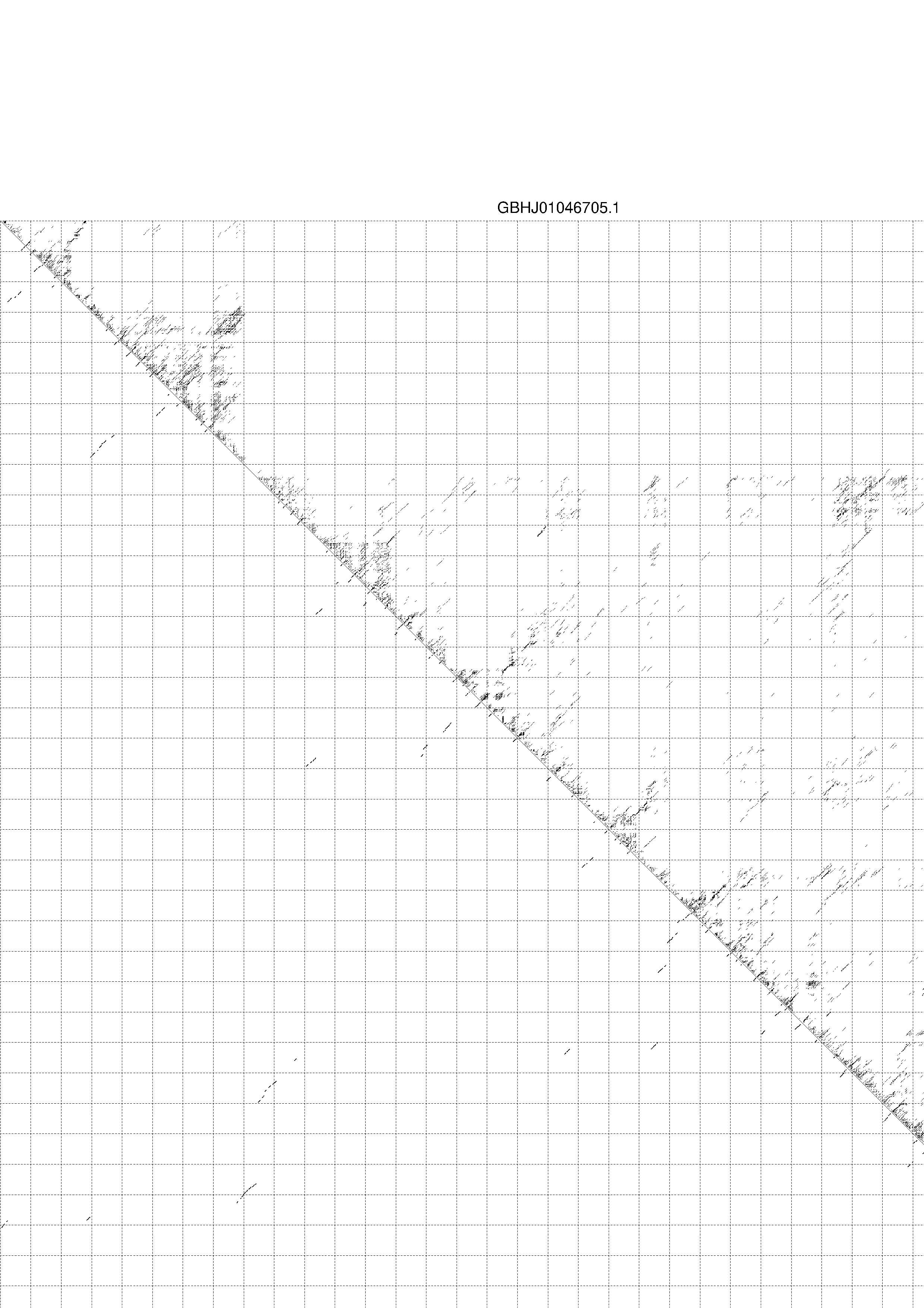
4. You may look at the dot plot containing the base pair probabilities [below]
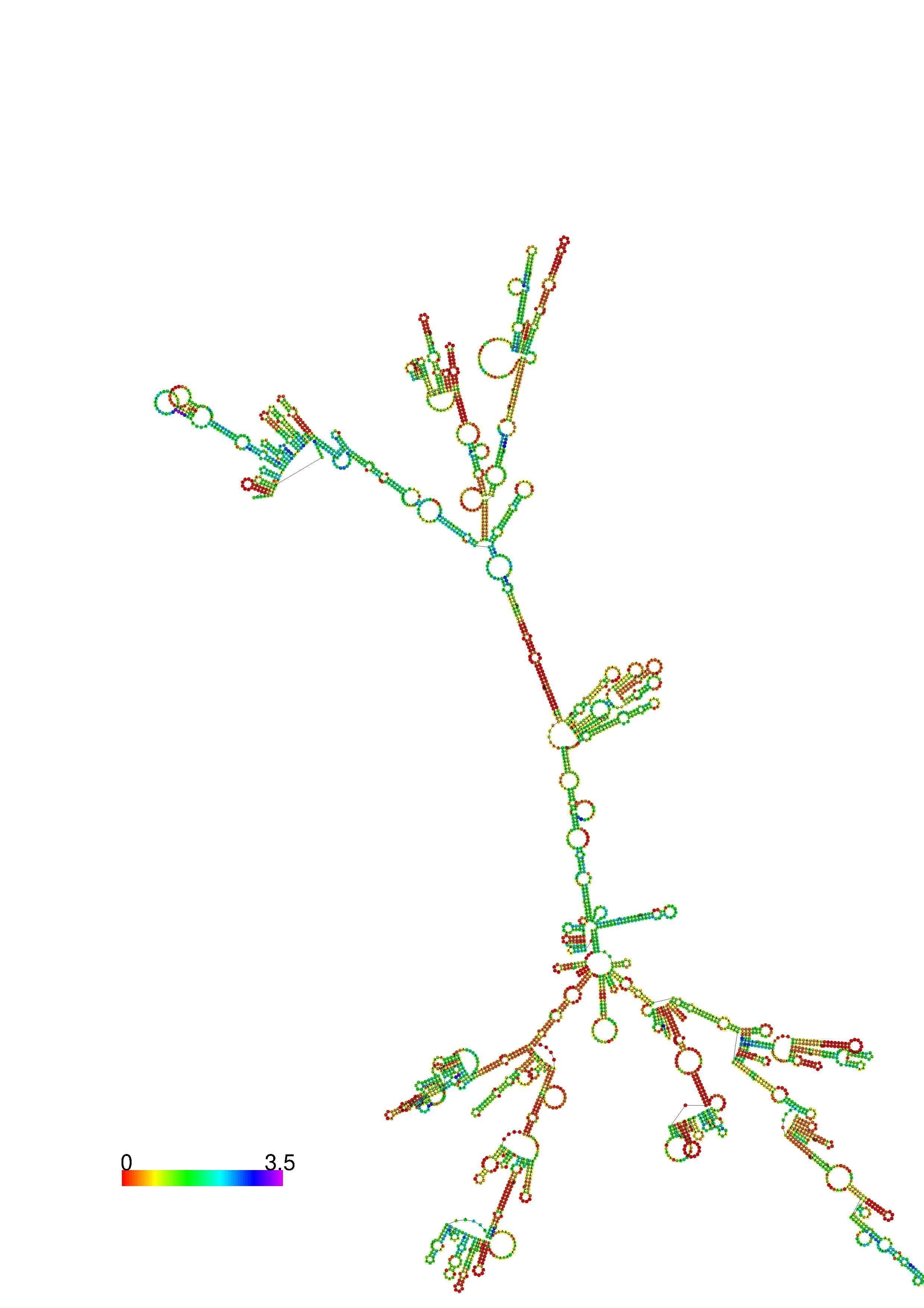
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
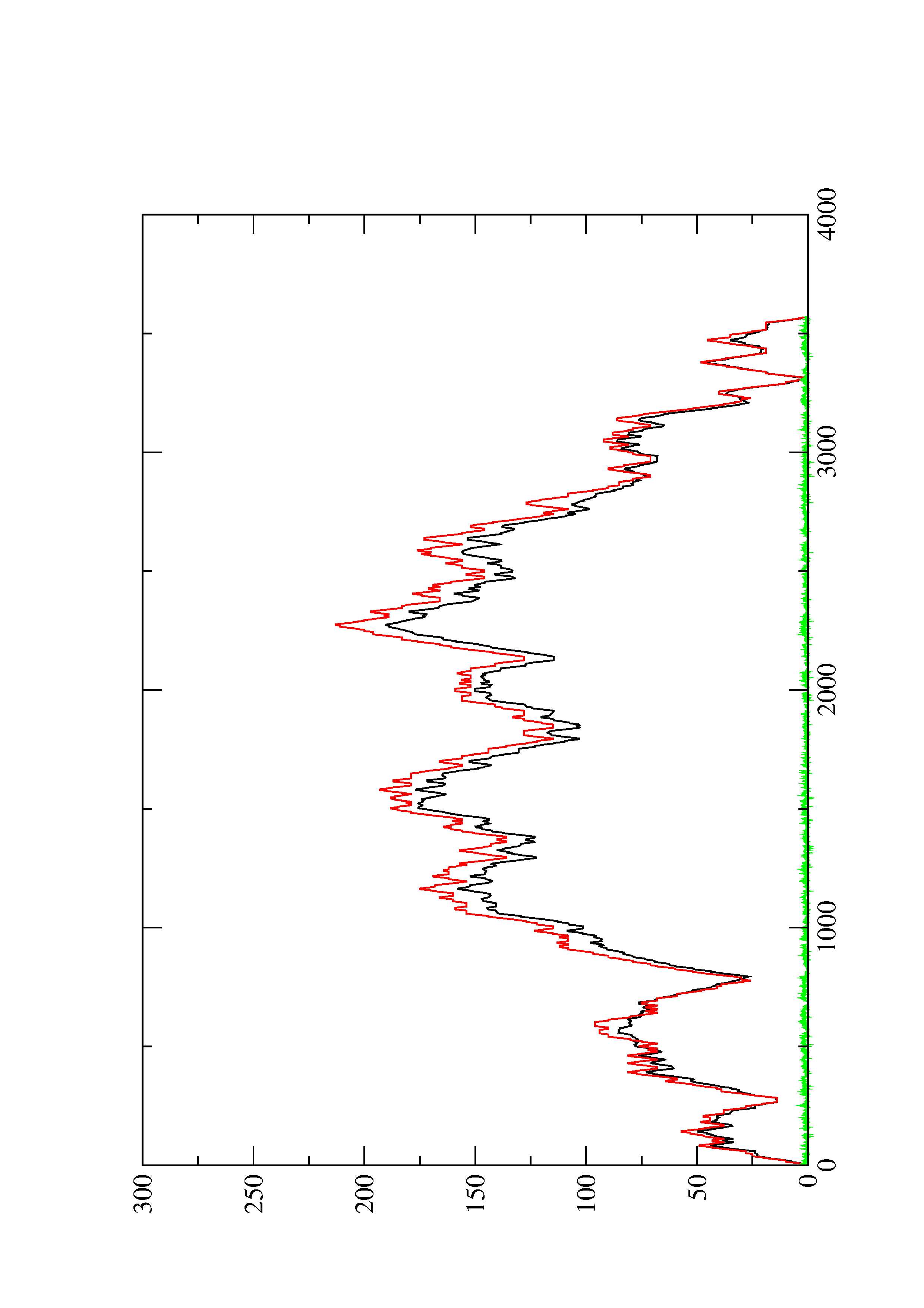
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.