lncRNA Structure Prediction
Species Name : Withania somnifera Sequence Id : GBHJ01046733.1
I. Result for minimum free energy prediction
The optimal secondary structure in dot-bracket notation with a minimum free energy of (-644.10) is given below.
1. Sequence-
GAUAAUAUCAUUUUAUUAUUGUAAAAAUUCGAGAAGGUGGGGUCCUCGUACACUGGGUUU
ACACUUGGAUUCCAUAAAUAACAAUACAACACAAAAUGAAUAAGAGAAAAUAAAACACAA
GAUAAAGAGGAUAAGAUAUCUUGGACCGUCGGGGCGUAAUUCUUAUGAGGCCAAUCAUAU
AUGGAUUUCGGACUCAUUGAAUAAGAGAGGUAUCACGUGGAAUCAUAUAUGGAUUUCGGA
CUCAUUGACUAAGAGAGGUAUCACCGUGGACAUGCAUAAGAUGAAGUAAAGUAUUAGUAU
CCCAUAAAAUAACUAAAGAACAAGUUAUGAAAAAUAAUGAAUGCUUACAUACAAGUUUAC
UAAUAUGAGUAGCCCAAACAAAACUAAGCCAAUCAUGGCCCAACAAGAGAAUCAAACAUC
AUCAUUAUUGUUAAGGCAAAAAGUAAAACACAGGGGUUUAUAAGAUGGCCAAGUGGCACA
UUAAACUUUGUCUCAAGGACCAUACACGAGUGAAGAUAGUGUACACAAGUUUAAUAGGAA
UUAACAUGCUGAAUUUGAUAAUAUCACAGUUCAAUUCAUCUAAACACGUAUAAAUCAUGA
UAAAGUCAUACUGAGAAGAAGGAACAUACACACUUCAAUGACAUCAAAAUUAACACUAUA
UUGCCUUAUAACCAUAUUAUAAGCCUAUUAAGCAACUUAAAACGGGAUUGCACAAAAACC
UCACUUACUUCAUGUAUCUUAGUAUCUAUUUCAUUAAAUAAAAAAUUCCACGAGUGAGUC
AUUCACGUAGCAAAGAGUGCUAGCACUAGGUCAUUUUAUAUUGAGUUUAUACAAUCCAGG
UUACUCUAGCUCAUGAUUUAAAGGGACAGUUUCAGCCAUAUAACAUACUGGAUUAAAGGU
AAAAUAGAUAGAAGAUGUGAUCAUAUUGUUGAUUAAAUCAGGUAACAUGCGUAAUUACAU
AGAAAAGACCCUAUUAUAUCCAUAAUAACACCUCAUAUACAAGAUAUUACAAGUAAGGUC
AUUUAAAAAUAAAGUAGACAGAUUCAUGUUCAGUCUGUGUAAUUGACUGUUAUCUUAUCA
UCAGGUCCAGGGUCAUAACAAGUAUUAGCAUCUUUAGGCAAACUCAGGUGUAAUUUAUCA
AGUAGAAACAUUCAAGUUUCCAUGCAAAGCCAUAUGGUCUACAAUACUUAUAAAGGAAUC
AUUUAACACAGUUUGUUGGAAACAGAUGCACGCAUUGGAGGGUAAAGACCAUAUACAAGA
AAGUUAAUUAAUCUAGAUUAAUGAGAACAGGGUACACAUGUUAGAGAAUUGUACAGGGUG
CACAUAUGUAGUUGAAUCAAAGGGAAAUCAUUAUUAUUCAACUGAGGAAUGGCACAUAUA
CAAGCAAGGUAUGGUCACACUCAAGCAUGUGAAAUUAUCAUGUUAUGGGGAGAAAGGUCU
GAGAGAGUAAAUAAGUGAUCACAUAUGACAAAGACAAUCAACCUUAUGACUUCAAUACUC
AAUUAAGUAGGCUAAUAUCAUCUCACUAGGCAAAACAUUCUUUUAACUAGUGGCAAGCAG
ACUUGGGGUCAUGCUUUCUAACCAACAGAAGCAGAUAGUGAGCUUAAGAUAGAGAUUGCA
AUCAAAUUCAAAUUCAGGAAGUAAGGCAACAUGGUUAGAAAAGGAAGUGAAUAACACAAC
ACUCUUGGUUUUUCAUCAAACUAGGUCUAAAUACUACCUAAAGGGAGAAUAUAGGAACUA
GGUCUAAAUACUACCUAAAGGGAGAAUACAGGACUCACACAAGAUCAGAUGAUGUGCAUC
CAACUAAUCAUCUAAAAACAAGACCAUUGGUACUUAUAUUUAUCUAUCUACAAGCUUUAC
UACUUCUUGGUUGCAGGAUUGCUAUUACAGUUAUAACAAAUAAGAAAAGCCAUCAUUACA
GUUUUCAAAGGGAGAAAUAACAAAUAAGAAAAGCCAUCAUUACAGUUUUCAACGGGAGAA
ACAGACAGGCUUAUAUACUUAAAAUUAUAGUAGUGCAGGCAGUAGGCAACCUAGUGAUGA
UUCUAUAUCAAAAGAUCCCAUUUUGACAAGAACUAAAGGGAAGAAAACAUACACAAGAAC
UUCAAUUAUGCUAGGCUCUCUAGUCAAUCUUCAAAUUCAUAGUUAAGCACAGAGUAUCAA
CAUGUAAGAGAGAGAAGGGGCAGACAACAAGGUUAUAAAAAUCAAGGACUAGGAGGAUGA
AUAGUUGAGGUAGGGAUUAUGAUAUAUCAUACUAACAUAAGUAGAAAGUGACCAAGACUA
ACUUAUAUUACAUACAAAUCAGGUUCUUAAUAAACAACUAACUUGGAAUGAGCUAAAAUG
ACCAAAAGUGGGUGGUAGUAGGAAACAACUUAGCCAAAUCAUACGCUUUAUCUUUUGAAG
UUGGACUUGUUGAAGGAAAAAUAUCAAACUACCCUUAUCACAUAUAUAGUUAAGGGACAA
GAGUAAUACGUUCACCAAUUUAUCUUUAAUAAAAUACAACAUAUGAGUAUUAUGCCAUUU
CAGUGGAAUCCUUUUGGCUCAAAAGUUAAAACAUUGAGGUUUAGUACAUUGCCCAUAAGG
AUGGAUGGAAAUAAACUCAUCAUUGACUCUUAAGGAAUCUACAAGUACCUGAAAGUAUUA
CAGGAUCAAGUAAUUUUUUCUUGGAUAAUGGACUAGUCAAUCAUGAGAAAUGCAAGCUGA
CAUUCAUAAGACCAAUGUACGUAGAUAACAUAGGCACAUGGUAAGUCAUAUUUGCAUGCA
UUUAGGCCUAACAUGAGAAGUUUCUAACUUUACUAACUCAGCCAACUAACCAUUACUUCA
AUUAGCGAUGAAUAGAUUUAAAUUCUCAUUUUCAAGACAAACAGAUCCACAAAACCAUAU
AUCAUAUAAGAUGGGGGUCAAAUGCAUGAAAAACAAGGAUAGAAUACAUAAACAAAGAGA
UAAAAUGUUCAAUUUCACUCAACCUUAGCCUAAAUAUCAUUUUUAAAAAUGGGUAAGGUC
ACACAAUCAACUAUAUUCAAGAAGGCAAUCAUAUCCUCUACACAUAUUUAGGUAUAAUCC
AUAUCAUCUCAAGAGAUGAAGUAGAUGUCAAUUUAUUAAAACUUUUAACAUCUUAAAGAA
ACUGUGUACUGGGAGAUCAUUGACUUCACAUAUACGCAUGUUGGCAUAAAAGAAAACAAG
AAGAAACAGUUAUGUUCAUGAGGUUACAUACAAAUGGGUGUAAUGUUCAAGUCUCAGGAG
GAUCAUUUACAAAUUAUAAGGCCAUUCUUAUGUGAAGAUUUAGACUUGUGUGAGAACUGA
AAAGAUUUACAAACAAGGAACAAGAUCUGAGUAGAACCGGGAAAUAAGUAUUUUUACCCU
UGCAAGAUUAAAAUAUUUCUUAACAACUAACAGCCUAUGGACUAAAUACAC
2. MFE structure-
(((((((......)))))))..........((((((((..((((((.(((.(((((((....(((((((((((.............((........))..((((((..(((......(((((((............)))))))..((....)).....(((((((((.(((....((((((((.((((((..(((.((..
...)).)))........))))))))))))))........((((.(((.(((...(((...((((((((((.((((.(((((((((((((.(((((((((.........((((((........))))))..............((((......)))).))))))))).....(((..(((((......((((....)))).
......((((........)).))....)))))..)))................((((((((((.((((......((((.......(((((.(((.............))))))))(((((((......(((((....((((((((.((.((((((....))).)))))))).)))))...)))))............(((
(...(((((........((((...........)))).)))))))))......)))))))..))))......)))).)))))))))).....(((((((.....))).)))).............))))))))....))))))))).(((((....)))))......)))))).))))....)))...)))))).))))((
(...(((..(((.(((((...((((((....................)))))).....))))).))))))...)))(((((..........((((((((((((..........(((.((((((......)))))).)))((((((((....((((((((((((...((((.(((((((((....(((........)))..
..)))))....)))).))))................((((......))))...))))))))))))..))))))))..(((.((((((((((.((((..(((((....((((((...((((((((...((((..(((((.((((((((((......)))))........(((....)))......))))))))))......
.....))))))))))))......)))))).....(((...(((....))).....)))....(((((((......)))))))((((((..(((....((((.......(((((....((((.(((...(((((((((..................)))))))))....)))))))...)))))....(((.(((((((.(
(((((((((.........)))))).))))......((((((...(((((...((((.((......(((.........))).....)).))))...))))).......))))))...(((((((((((((..((((.....))))..))))))).(((((((((...)))).)))))..((((((..(((((....(((((
(((((.((((((((((....)))............(((.((((((((....((((.(((((((((.(((..........)))))))..))))).))))......))))...)))).))).......(((((((((.(((((((((.....((.(((...)))))......(((..((((((........)).))))..))
)...................)))))..)))).))))))))).)))))))..)))).))))))))))))))))).(((.(((((..(((..((((.....)))).....(((..((((((..(((((.....)))))..............(((((..........(((((.....))))).......)))))........
.........))))))..))).)))))))))))...))))))..(((((((...((((((.((((....))))..........................(((((((((((......(((.((((((..(((((...(((((((((..((.....))...))).)))..)))...)))))............((((.....)
)))...))))))))).....))))))))))).)))))).)))))))((.((((......))))))..)))))))..)))..........))))....)))..))))))............)))))..)))).)).............((((.((((..((((......))))))))..))))..((((((.....)))))
))))))))).)))..................((((((.............))))))......................))))))))))))..........))))).((((((.(((...((((((.....((((((...))))))......))))))))).))))))...)))))))))))).(((((........))))
))))..)))))).))))))..)))))))))...)))..))))))))).....))))))))((((...(((((...((((..(((.....))).....)))).)))))....))))((((..(((....(((((((...(((((((..((((((.(((((((((.(((((....((((.(((..........)))..))))
..........((((.((......)).))))....(((((....(((........))).....))))).......((((.(((...)))..)))))))))))))))))).....(((((..(((((((.........(((...((((.....))))..))).((((((.((....................)).)))))).
.................................((((.......(((((((((....(((...((((((....))))))(((((((....)))))))....(((((....((((.....(((((((((.((((((........)))).))..)))))).))).......)))).....))))).((((....))))...(
(.(((((((........))))))).))(((((((.....(((((.((((((.....(((((....)))))))))))))))))))))))((((((..((....)).)))))).....))).....))))))))).....)))).....)))))))...)))))........))))))..)))....))))...))))))).
.)))..)))).
You can download the minimum free energy (MFE) structure in
II. Results for thermodynamic ensemble prediction
1. The free energy of the thermodynamic ensemble is [-712.47] kcal/mol.
2. The frequency of mfe structure in ensemble 6.64546e-49.
3. The ensemble diversity 803.15.
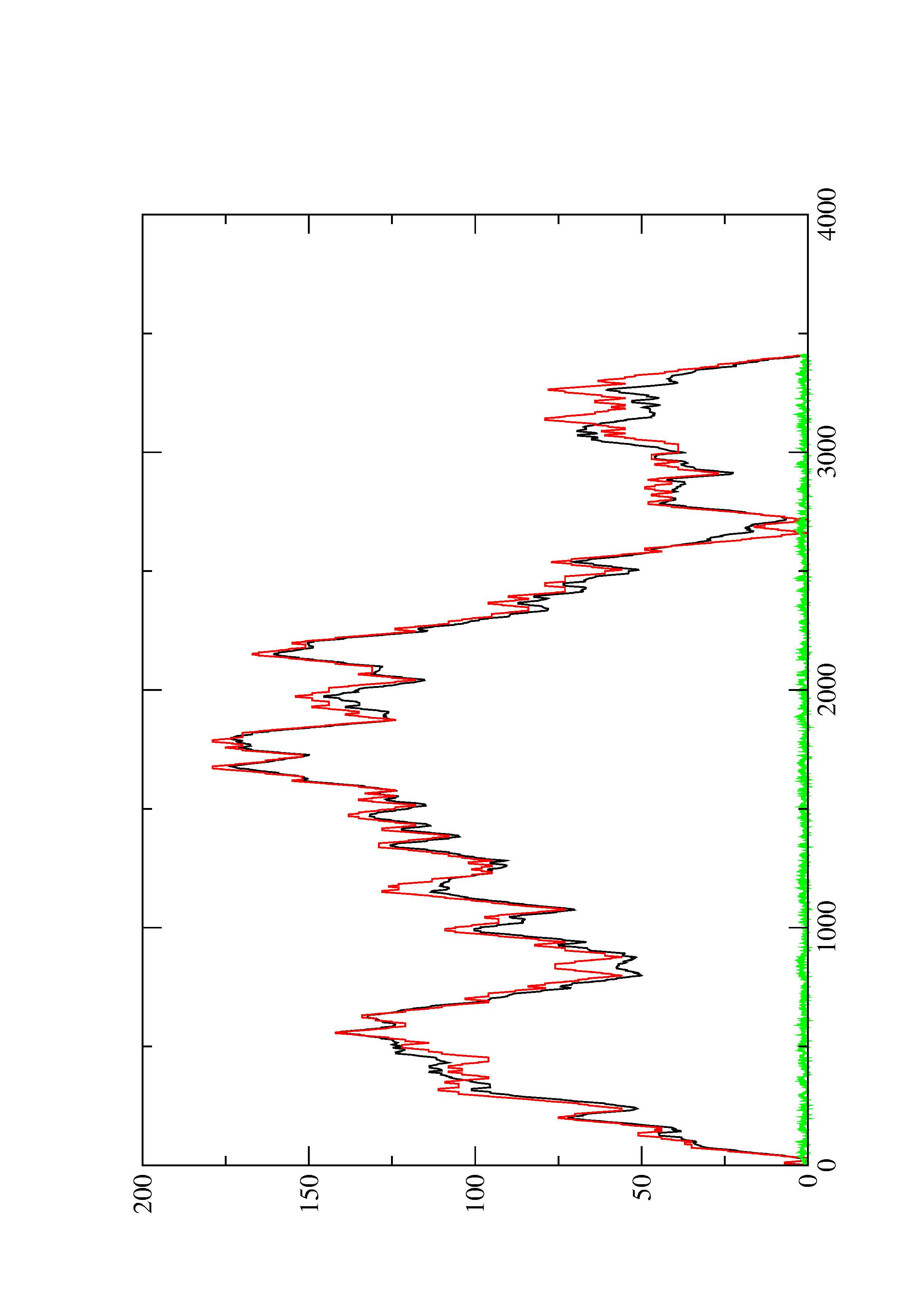
4. You may look at the dot plot containing the base pair probabilities [below]
III. Graphical output
1. A note on base-pairing probabilities:--
The structure below is colored by base-pairing probabilities. For unpaired regions the color denotes the probability of being unpaired. The reliability information of RNA secondary structure
plot in the form of color annotation, Positional entropy ranging from red (low entropy, well-defined) via green to blue and
violet (high entropy, ill-defined).
2. Here you find a mountain plot representation of the MFE structure, the thermodynamic ensemble of RNA structures, and the centroid structure. Additionally we present the positional entropy for each position. The resulting plot shows three curves, two mountain plots derived from the MFE structure (red) and the pairing probabilities (black) and a positional entropy curve (green). Well-defined regions are identified by low entropy.