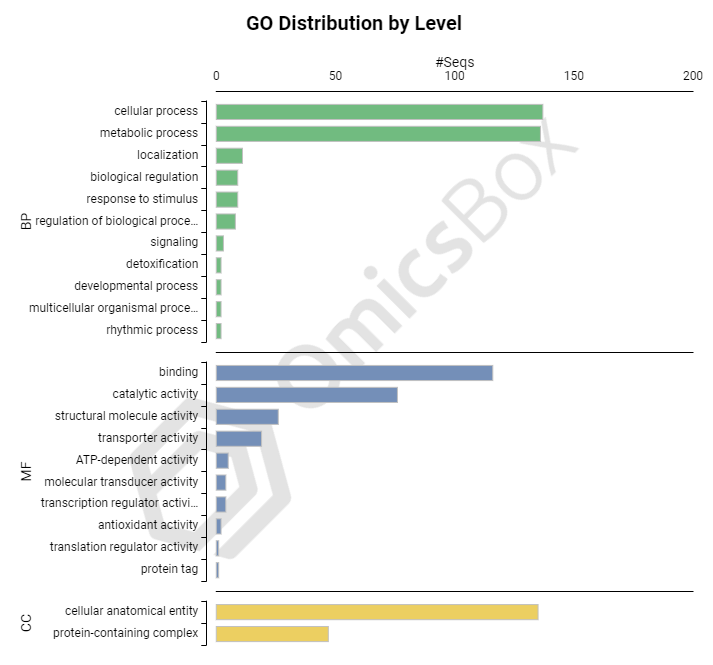
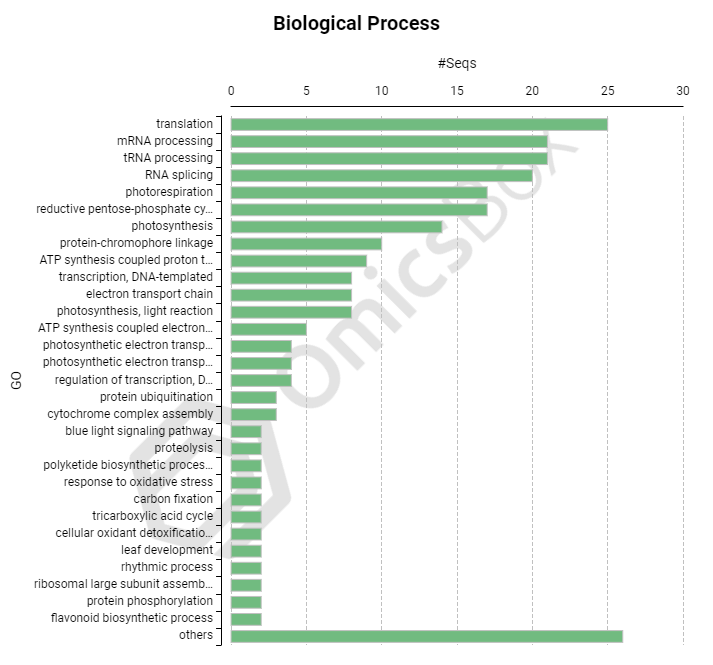
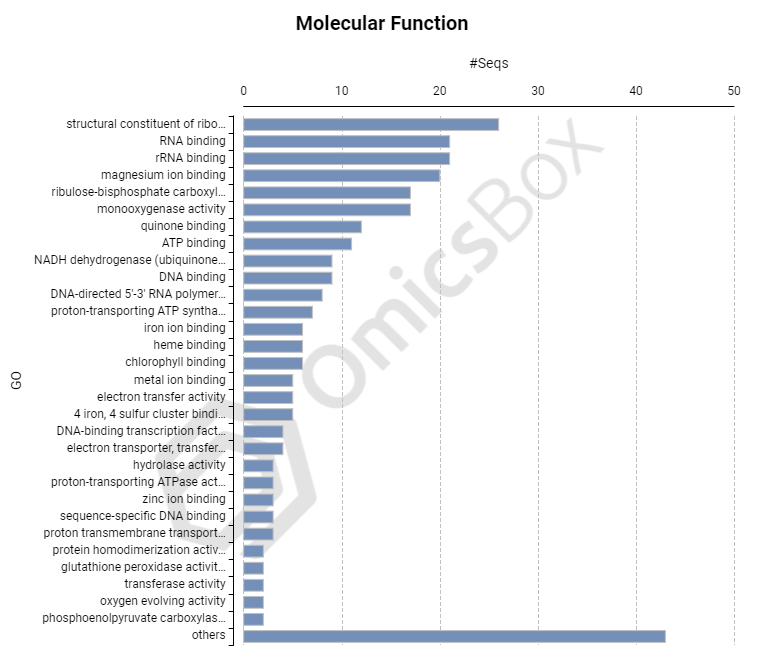
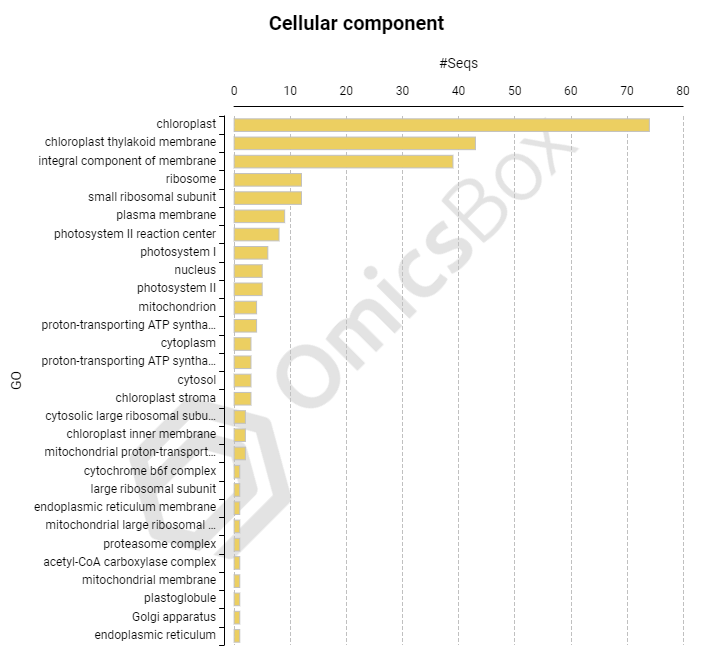
Coding Information
| Acc No. | Species Name | Description | Length | GO IDs | GO Names | Enzyme Names | InterPro GO Names | Peptide Information | BLAST | Nucleotide Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| KY536305.1 | Acorus calamus | WD repeat-containing protein 43 isoform X1 | 150 | P:GO:0006364; C:GO:0005730 | P:rRNA processing; C:nucleolus | P:regulation of transcription, DNA-templated | view details | |||
| KY530996.1 | Acorus calamus | TIMELESS-interacting protein isoform X1 | 213 | P:GO:0000076; P:GO:0006974; P:GO:0043111; P:GO:0048478; F:GO:0003677; F:GO:0008270; C:GO:0016021; C:GO:0031298 | P:DNA replication checkpoint signaling; P:cellular response to DNA damage stimulus; P:replication fork arrest; P:replication fork protection; F:DNA binding; F:zinc ion binding; C:integral component of membrane; C:replication fork protection complex | P:DNA replication checkpoint signaling; P:cellular response to DNA damage stimulus; P:replication fork protection; C:nucleus | view details | |||
| MF694654.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM398149.1 | Acorus calamus | PHP | 948 | P:GO:0006368; P:GO:0016570; C:GO:0016593 | P:transcription elongation from RNA polymerase II promoter; P:histone modification; C:Cdc73/Paf1 complex | P:transcription elongation from RNA polymerase II promoter; P:histone modification; C:Cdc73/Paf1 complex | view details | |||
| KM399340.1 | Acorus calamus | tunicamycin induced 1 | 915 | C:GO:0005783 | C:endoplasmic reticulum | no GO terms | view details | |||
| HO002070.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY533969.1 | Acorus calamus | pentatricopeptide repeat-containing protein At1g08610-like | 1383 | F:protein binding | view details | |||||
| KY527118.1 | Acorus calamus | putative nitric oxide synthase isoform X1 | 1500 | F:GO:0003924; F:GO:0005525; C:GO:0009507 | F:GTPase activity; F:GTP binding; C:chloroplast | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| KY538987.1 | Acorus calamus | pentatricopeptide repeat-containing protein At4g20740 | 1902 | F:protein binding | view details | |||||
| AF197621.1 | Acorus calamus | ATPase subunit 1 | 1240 | P:GO:0015986; F:GO:0005524; F:GO:0043531; F:GO:0046933; C:GO:0005743; C:GO:0045261 | P:ATP synthesis coupled proton transport; F:ATP binding; F:ADP binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:mitochondrial inner membrane; C:proton-transporting ATP synthase complex, catalytic core F(1) | H(+)-transporting two-sector ATPase; Ligases | no IPS match | view details | ||
| AM889577.1 | Acorus calamus | NADH dehydrogenase subunit J | 431 | P:GO:0019684; P:GO:0022900; F:GO:0008137; F:GO:0048038; C:GO:0005743; C:GO:0005886; C:GO:0009535 | P:photosynthesis, light reaction; P:electron transport chain; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:mitochondrial inner membrane; C:plasma membrane; C:chloroplast thylakoid membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| KY536094.1 | Acorus calamus | inhibitor of Bruton tyrosine kinase | 3045 | P:GO:0016310; F:GO:0016301 | P:phosphorylation; F:kinase activity | F:protein binding | view details | |||
| KY538498.1 | Acorus calamus | UNC93-like protein 3 | 1215 | P:GO:0055075; C:GO:0005886; C:GO:0016021 | P:potassium ion homeostasis; C:plasma membrane; C:integral component of membrane | P:potassium ion homeostasis | view details | |||
| KM400265.1 | Acorus calamus | ARM repeat superfamily protein | 1380 | P:GO:0043248 | P:proteasome assembly | P:proteasome assembly | view details | |||
| FJ875015.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MN481071.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY539040.1 | Acorus calamus | probable thimet oligopeptidase isoform X1 | 1635 | P:GO:0006508; P:GO:0006518; F:GO:0004222; F:GO:0046872; C:GO:0016021 | P:proteolysis; P:peptide metabolic process; F:metalloendopeptidase activity; F:metal ion binding; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:metalloendopeptidase activity; F:metallopeptidase activity | view details | ||
| KM399174.1 | Acorus calamus | G-protein coupled receptor 1 | 888 | P:GO:0000278; P:GO:0006571; P:GO:0007166; P:GO:0007189; P:GO:0007202; P:GO:0009094; P:GO:0009735; P:GO:0009742; P:GO:0009785; P:GO:0009788; P:GO:0009908; P:GO:0009939; P:GO:0010231; P:GO:0010244; P:GO:0032960; F:GO:0004930; C:GO:0005886; C:GO:0016021 | P:mitotic cell cycle; P:tyrosine biosynthetic process; P:cell surface receptor signaling pathway; P:adenylate cyclase-activating G protein-coupled receptor signaling pathway; P:activation of phospholipase C activity; P:L-phenylalanine biosynthetic process; P:response to cytokinin; P:brassinosteroid mediated signaling pathway; P:blue light signaling pathway; P:negative regulation of abscisic acid-activated signaling pathway; P:flower development; P:positive regulation of gibberellic acid mediated signaling pathway; P:maintenance of seed dormancy; P:response to low fluence blue light stimulus by blue low-fluence system; P:regulation of inositol trisphosphate biosynthetic process; F:G protein-coupled receptor activity; C:plasma membrane; C:integral component of membrane | P:cell surface receptor signaling pathway; F:transmembrane signaling receptor activity; C:integral component of membrane | view details | |||
| KM400372.1 | Acorus calamus | arginine-rich cyclin 1 | 525 | P:GO:0006357; P:GO:0007049; P:GO:0050790; P:GO:0051301; F:GO:0016538 | P:regulation of transcription by RNA polymerase II; P:cell cycle; P:regulation of catalytic activity; P:cell division; F:cyclin-dependent protein serine/threonine kinase regulator activity | P:regulation of transcription by RNA polymerase II; F:cyclin-dependent protein serine/threonine kinase regulator activity | view details | |||
| KY529019.1 | Acorus calamus | DNA damage-binding protein 2 | 1422 | P:GO:0006281; P:GO:0010224; F:GO:0003684; C:GO:0005634; C:GO:0080008 | P:DNA repair; P:response to UV-B; F:damaged DNA binding; C:nucleus; C:Cul4-RING E3 ubiquitin ligase complex | P:DNA repair; F:damaged DNA binding; F:protein binding; C:nucleus; C:Cul4-RING E3 ubiquitin ligase complex | view details | |||
| HO002074.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| DQ859124.1 | Acorus calamus | apocytochrome b | 1023 | P:GO:0022904; P:GO:1902600; F:GO:0008121; F:GO:0046872; C:GO:0005743; C:GO:0045275 | P:respiratory electron transport chain; P:proton transmembrane transport; F:ubiquinol-cytochrome-c reductase activity; F:metal ion binding; C:mitochondrial inner membrane; C:respiratory chain complex III | Acting on diphenols and related substances as donors; Quinol--cytochrome-c reductase | P:respiratory electron transport chain; F:ubiquinol-cytochrome-c reductase activity; F:electron transfer activity; F:oxidoreductase activity; C:membrane; C:respiratory chain complex III | view details | ||
| KY538694.1 | Acorus calamus | probable zinc metalloprotease EGY2, chloroplastic isoform X2 | 1113 | P:GO:0006508; F:GO:0008237; C:GO:0016021; C:GO:0031969 | P:proteolysis; F:metallopeptidase activity; C:integral component of membrane; C:chloroplast membrane | Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| KY532556.1 | Acorus calamus | meiotic recombination protein SPO11-1 | 456 | P:GO:0000706; P:GO:0007131; P:GO:0042138; F:GO:0003677; F:GO:0003918; F:GO:0005524; F:GO:0016787; F:GO:0046872; C:GO:0000228 | P:meiotic DNA double-strand break processing; P:reciprocal meiotic recombination; P:meiotic DNA double-strand break formation; F:DNA binding; F:DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity; F:ATP binding; F:hydrolase activity; F:metal ion binding; C:nuclear chromosome | Hydrolases; DNA topoisomerase (ATP-hydrolyzing) | P:meiotic DNA double-strand break formation; F:DNA binding; F:DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity; C:chromosome | view details | ||
| KY527655.1 | Acorus calamus | putative methyltransferase NSUN6 isoform X2 | 1320 | P:GO:0001510; F:GO:0003723; F:GO:0008168 | P:RNA methylation; F:RNA binding; F:methyltransferase activity | Transferring one-carbon groups | P:RNA methylation; F:RNA binding; F:methyltransferase activity | view details | ||
| KY533214.1 | Acorus calamus | DNA replication licensing factor MCM4 | 1659 | P:GO:0000727; P:GO:0006268; P:GO:0006271; P:GO:0009555; P:GO:1902600; P:GO:1902975; F:GO:0003697; F:GO:0005524; F:GO:0015078; F:GO:0016887; F:GO:0017116; C:GO:0000347; C:GO:0000785; C:GO:0009507; C:GO:0016021; C:GO:0033179; C:GO:0042555 | P:double-strand break repair via break-induced replication; P:DNA unwinding involved in DNA replication; P:DNA strand elongation involved in DNA replication; P:pollen development; P:proton transmembrane transport; P:mitotic DNA replication initiation; F:single-stranded DNA binding; F:ATP binding; F:proton transmembrane transporter activity; F:ATP hydrolysis activity; F:single-stranded DNA helicase activity; C:THO complex; C:chromatin; C:chloroplast; C:integral component of membrane; C:proton-transporting V-type ATPase, V0 domain; C:MCM complex | Acting on acid anhydrides; DNA helicase; Translocases | P:DNA replication initiation; P:DNA duplex unwinding; F:DNA binding; F:DNA helicase activity; F:ATP binding; C:MCM complex | view details | ||
| KY525562.1 | Acorus calamus | transcription termination factor MTERF2, chloroplastic isoform X1 | 1284 | P:GO:0006353; P:GO:0006355; P:GO:0032502; F:GO:0003690; C:GO:0009507 | P:DNA-templated transcription, termination; P:regulation of transcription, DNA-templated; P:developmental process; F:double-stranded DNA binding; C:chloroplast | P:regulation of transcription, DNA-templated; F:double-stranded DNA binding | view details | |||
| MN481068.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY526680.1 | Acorus calamus | BRAP2 RING ZnF UBP domain-containing protein 2 | 1377 | P:GO:0044238; P:GO:0044260; F:GO:0008270 | P:primary metabolic process; P:cellular macromolecule metabolic process; F:zinc ion binding | F:zinc ion binding | view details | |||
| AM889964.1 | Acorus calamus | ycf2 protein | 353 | P:GO:0050790; F:GO:0005096; F:GO:0005524; F:GO:0016887; C:GO:0009570; C:GO:0016021 | P:regulation of catalytic activity; F:GTPase activator activity; F:ATP binding; F:ATP hydrolysis activity; C:chloroplast stroma; C:integral component of membrane | Acting on acid anhydrides | no GO terms | view details | ||
| KY524993.1 | Acorus calamus | anaphase-promoting complex subunit 2 isoform X1 | 2241 | P:GO:0009987; C:GO:0005680 | P:cellular process; C:anaphase-promoting complex | P:ubiquitin-dependent protein catabolic process; F:ubiquitin protein ligase binding | view details | |||
| KY535082.1 | Acorus calamus | Aspartate/glutamate/uridylate kinase | 855 | P:GO:0006225; P:GO:0016310; P:GO:0046940; F:GO:0033862; C:GO:0005829 | P:UDP biosynthetic process; P:phosphorylation; P:nucleoside monophosphate phosphorylation; F:UMP kinase activity; C:cytosol | Nucleoside-phosphate kinase; UMP kinase | P:pyrimidine nucleotide biosynthetic process; F:uridylate kinase activity; F:UMP kinase activity; C:cytoplasm | view details | ||
| FM882136.1 | Acorus calamus | WUSCHEL-related homeobox 7-like | 152 | P:GO:0006355; P:GO:0099402; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| JN090099.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| D28865.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1263 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM397607.1 | Acorus calamus | minichromosome maintenance 5 protein | 969 | P:GO:0006270; P:GO:0007049; P:GO:0032508; F:GO:0003678; F:GO:0003688; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0009507; C:GO:0042555 | P:DNA replication initiation; P:cell cycle; P:DNA duplex unwinding; F:DNA helicase activity; F:DNA replication origin binding; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:chloroplast; C:MCM complex | Acting on acid anhydrides; DNA helicase | P:DNA replication initiation; P:DNA duplex unwinding; F:DNA binding; F:DNA replication origin binding; F:ATP binding; C:nucleus; C:MCM complex | view details | ||
| AM889982.1 | Acorus calamus | cytochrome c heme attachment protein | 350 | P:GO:0017004; F:GO:0020037; C:GO:0009535; C:GO:0016021 | P:cytochrome complex assembly; F:heme binding; C:chloroplast thylakoid membrane; C:integral component of membrane | P:cytochrome complex assembly; F:heme binding | view details | |||
| KM399773.1 | Acorus calamus | probable polygalacturonase | 963 | P:GO:0005975; F:GO:0004650; F:GO:0016829 | P:carbohydrate metabolic process; F:polygalacturonase activity; F:lyase activity | Lyases; Endo-polygalacturonase | P:carbohydrate metabolic process; F:polygalacturonase activity | view details | ||
| KM398531.1 | Acorus calamus | annexin ANNAT3 | 828 | P:GO:0006950; F:GO:0005509; F:GO:0005544 | P:response to stress; F:calcium ion binding; F:calcium-dependent phospholipid binding | F:calcium ion binding; F:calcium-dependent phospholipid binding | view details | |||
| HO002069.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY540419.1 | Acorus calamus | fatty acid amide hydrolase | 1818 | P:GO:0042742; P:GO:0070291; F:GO:0016740; F:GO:0047412; F:GO:0050566; F:GO:0050567; C:GO:0005886; C:GO:0016021 | P:defense response to bacterium; P:N-acylethanolamine metabolic process; F:transferase activity; F:N-(long-chain-acyl)ethanolamine deacylase activity; F:asparaginyl-tRNA synthase (glutamine-hydrolyzing) activity; F:glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity; C:plasma membrane; C:integral component of membrane | Asparaginyl-tRNA synthase (glutamine-hydrolyzing); Glutaminyl-tRNA synthase (glutamine-hydrolyzing); Transferases; N-(long-chain-acyl)ethanolamine deacylase | F:catalytic activity | view details | ||
| KY538778.1 | Acorus calamus | PAN domain-containing protein At5g03700-like | 909 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| KY540093.1 | Acorus calamus | probable zinc metalloprotease EGY1, chloroplastic | 1629 | P:GO:0006508; P:GO:0009416; P:GO:0009658; P:GO:0009723; P:GO:0009959; P:GO:0010027; P:GO:0010207; P:GO:0043157; P:GO:0048564; P:GO:0060359; F:GO:0004222; C:GO:0016021; C:GO:0031969 | P:proteolysis; P:response to light stimulus; P:chloroplast organization; P:response to ethylene; P:negative gravitropism; P:thylakoid membrane organization; P:photosystem II assembly; P:response to cation stress; P:photosystem I assembly; P:response to ammonium ion; F:metalloendopeptidase activity; C:integral component of membrane; C:chloroplast membrane | Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| MN481075.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JF953047.1 | Acorus calamus | maturase K | 714 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KY525445.1 | Acorus calamus | protein SEEDLING PLASTID DEVELOPMENT 1 | 1803 | P:GO:0009657; P:GO:0010431; P:GO:0090677; F:GO:0005524; F:GO:0016887; C:GO:0031972 | P:plastid organization; P:seed maturation; P:reversible differentiation; F:ATP binding; F:ATP hydrolysis activity; C:chloroplast intermembrane space | Acting on acid anhydrides | no GO terms | view details | ||
| KY524902.1 | Acorus calamus | thioredoxin-like fold domain-containing protein MRL7, chloroplastic | 582 | P:GO:0005975; P:GO:0006355; P:GO:0009658; F:GO:0004553; C:GO:0042644 | P:carbohydrate metabolic process; P:regulation of transcription, DNA-templated; P:chloroplast organization; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:chloroplast nucleoid | Glycosylases | P:regulation of transcription, DNA-templated; P:chloroplast organization | view details | ||
| MN216564.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 561 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY536198.1 | Acorus calamus | protein root UVB sensitive 5 | 1353 | no GO terms | view details | |||||
| AJ347846.1 | Acorus calamus | photosystem II CP47 chlorophyll apoprotein | 1361 | P:GO:0009772; P:GO:0018298; F:GO:0016168; F:GO:0045156; C:GO:0009523; C:GO:0009535; C:GO:0016021 | P:photosynthetic electron transport in photosystem II; P:protein-chromophore linkage; F:chlorophyll binding; F:electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity; C:photosystem II; C:chloroplast thylakoid membrane; C:integral component of membrane | Oxidoreductases | P:photosynthetic electron transport chain; P:photosynthetic electron transport in photosystem II; P:photosynthesis; P:photosynthesis, light reaction; F:chlorophyll binding; F:electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity; C:photosystem; C:photosystem II; C:membrane | view details | ||
| MF694941.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 553 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY534287.1 | Acorus calamus | ribosome recycling factor | 543 | P:GO:0006412; F:GO:0043023; C:GO:0005739 | P:translation; F:ribosomal large subunit binding; C:mitochondrion | P:translation | view details | |||
| KM400540.1 | Acorus calamus | regulatory particle non-ATPase 13 | 930 | P:GO:0006511; P:GO:0010950; F:GO:0043130; F:GO:0061133; F:GO:0070628; C:GO:0005634; C:GO:0005737; C:GO:0008541 | P:ubiquitin-dependent protein catabolic process; P:positive regulation of endopeptidase activity; F:ubiquitin binding; F:endopeptidase activator activity; F:proteasome binding; C:nucleus; C:cytoplasm; C:proteasome regulatory particle, lid subcomplex | C:nucleus; C:cytoplasm | view details | |||
| JF940685.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 615 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY539422.1 | Acorus calamus | riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 | 1083 | P:GO:0009231; P:GO:0009644; P:GO:0009658; P:GO:0046443; P:GO:1901135; F:GO:0008703; F:GO:0008835; F:GO:0016799; F:GO:0050661; C:GO:0009507 | P:riboflavin biosynthetic process; P:response to high light intensity; P:chloroplast organization; P:FAD metabolic process; P:carbohydrate derivative metabolic process; F:5-amino-6-(5-phosphoribosylamino)uracil reductase activity; F:diaminohydroxyphosphoribosylaminopyrimidine deaminase activity; F:hydrolase activity, hydrolyzing N-glycosyl compounds; F:NADP binding; C:chloroplast | Diaminohydroxyphosphoribosylaminopyrimidine deaminase; Glycosylases; 5-amino-6-(5-phosphoribosylamino)uracil reductase | P:riboflavin biosynthetic process; F:5-amino-6-(5-phosphoribosylamino)uracil reductase activity | view details | ||
| JN090015.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM399935.1 | Acorus calamus | accelerated cell death 11 | 591 | P:GO:0120009; F:GO:0120013; C:GO:0005737 | P:intermembrane lipid transfer; F:lipid transfer activity; C:cytoplasm | Translocases | P:intermembrane lipid transfer; F:lipid transfer activity; C:cytoplasm | view details | ||
| KM398963.1 | Acorus calamus | ABH1 | 2265 | P:GO:0000380; P:GO:0000394; P:GO:0031053; P:GO:0045292; P:GO:0048574; P:GO:0051028; P:GO:0051607; P:GO:1901527; F:GO:0000339; C:GO:0005634; C:GO:0005845; C:GO:0005846 | P:alternative mRNA splicing, via spliceosome; P:RNA splicing, via endonucleolytic cleavage and ligation; P:primary miRNA processing; P:mRNA cis splicing, via spliceosome; P:long-day photoperiodism, flowering; P:mRNA transport; P:defense response to virus; P:abscisic acid-activated signaling pathway involved in stomatal movement; F:RNA cap binding; C:nucleus; C:mRNA cap binding complex; C:nuclear cap binding complex | P:mRNA export from nucleus; P:RNA metabolic process; F:RNA cap binding; F:RNA binding; F:protein binding; C:nuclear cap binding complex | view details | |||
| FM882137.1 | Acorus calamus | wuschel related homeobox 4 | 158 | P:GO:0006355; P:GO:0010067; P:GO:0010087; P:GO:0051301; P:GO:0099402; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:procambium histogenesis; P:phloem or xylem histogenesis; P:cell division; P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| LC219274.1 | Acorus calamus | maturase K | 789 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KY533106.1 | Acorus calamus | dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase | 1587 | P:GO:0006487; P:GO:0097502; F:GO:0052918; F:GO:0052926; C:GO:0005789; C:GO:0016021 | P:protein N-linked glycosylation; P:mannosylation; F:dol-P-Man:Man(8)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase activity; F:dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | Dolichyl-P-Man:Man(8)GlcNAc(2)-PP-dolichol alpha-1,2-mannosyltransferase; Dolichyl-P-Man:Man(6)GlcNAc(2)-PP-dolichol alpha-1,2-mannosyltransferase | F:mannosyltransferase activity; F:glycosyltransferase activity | view details | ||
| DQ008879.1 | Acorus calamus | NADH dehydrogenase subunit F | 1998 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| GQ436696.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 703 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY531905.1 | Acorus calamus | A-kinase anchor protein 17A isoform X1 | 870 | P:GO:0016310; F:GO:0016301 | P:phosphorylation; F:kinase activity | no GO terms | view details | |||
| MZ190981.1 | Acorus calamus | hypothetical protein BVRB_036970, partial | 507 | no GO terms | view details | |||||
| KY526570.1 | Acorus calamus | thylakoid lumenal 15.0 kDa protein 2, chloroplastic | 558 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| AF195652.1 | Acorus calamus | ribosomal protein S2 | 273 | C:GO:0005739; C:GO:0005840 | C:mitochondrion; C:ribosome | no GO terms | view details | |||
| LC219032.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM398478.1 | Acorus calamus | diphthamide synthesis DPH2 family protein | 1023 | P:GO:0017183; F:GO:0090560 | P:peptidyl-diphthamide biosynthetic process from peptidyl-histidine; F:2-(3-amino-3-carboxypropyl)histidine synthase activity | 2-(3-amino-3-carboxypropyl)histidine synthase | P:peptidyl-diphthamide biosynthetic process from peptidyl-histidine | view details | ||
| KM399554.1 | Acorus calamus | haloacid dehalogenase-like hydrolase superfamily protein | 885 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| KY532409.1 | Acorus calamus | histone-lysine N-methyltransferase ASHH1 | 609 | P:GO:0006355; P:GO:0010452; F:GO:0046975; C:GO:0000785; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:histone H3-K36 methylation; F:histone methyltransferase activity (H3-K36 specific); C:chromatin; C:nucleus | [Histone H3]-lysine(4) N-trimethyltransferase | F:protein binding | view details | ||
| KY540747.1 | Acorus calamus | nuclear cap-binding protein subunit 1 | 2574 | P:GO:0000184; P:GO:0000380; P:GO:0000394; P:GO:0031053; P:GO:0045292; P:GO:0048574; P:GO:0051028; P:GO:0051607; P:GO:1901527; F:GO:0000339; F:GO:0003729; C:GO:0005634; C:GO:0005845; C:GO:0005846 | P:nuclear-transcribed mRNA catabolic process, nonsense-mediated decay; P:alternative mRNA splicing, via spliceosome; P:RNA splicing, via endonucleolytic cleavage and ligation; P:primary miRNA processing; P:mRNA cis splicing, via spliceosome; P:long-day photoperiodism, flowering; P:mRNA transport; P:defense response to virus; P:abscisic acid-activated signaling pathway involved in stomatal movement; F:RNA cap binding; F:mRNA binding; C:nucleus; C:mRNA cap binding complex; C:nuclear cap binding complex | P:mRNA export from nucleus; P:RNA metabolic process; F:RNA cap binding; F:RNA binding; F:protein binding; C:nuclear cap binding complex | view details | |||
| KM398852.1 | Acorus calamus | AT5G11810-like protein | 687 | no GO terms | view details | |||||
| KY541728.1 | Acorus calamus | protein CHAPERONE-LIKE PROTEIN OF POR1, chloroplastic | 597 | P:GO:0006457; P:GO:0008219; P:GO:0009658; P:GO:0009704; P:GO:0009793; P:GO:1904216; F:GO:0044183; C:GO:0005739; C:GO:0009534; C:GO:0009570; C:GO:0009706; C:GO:0016021; C:GO:0055035 | P:protein folding; P:cell death; P:chloroplast organization; P:de-etiolation; P:embryo development ending in seed dormancy; P:positive regulation of protein import into chloroplast stroma; F:protein folding chaperone; C:mitochondrion; C:chloroplast thylakoid; C:chloroplast stroma; C:chloroplast inner membrane; C:integral component of membrane; C:plastid thylakoid membrane | no GO terms | view details | |||
| LC219293.1 | Acorus calamus | maturase K | 786 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KM360612.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1345 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM823797.1 | Acorus calamus | rhomboid-like protein 14, mitochondrial | 999 | P:GO:0006508; F:GO:0004252; F:GO:0046872; C:GO:0016021 | P:proteolysis; F:serine-type endopeptidase activity; F:metal ion binding; C:integral component of membrane | Acting on peptide bonds (peptidases) | F:serine-type endopeptidase activity; C:integral component of membrane | view details | ||
| KU127422.1 | Acorus calamus | acetyl-CoA carboxylase carboxyltransferase beta subunit | 511 | P:GO:0006633; P:GO:2001295; F:GO:0003989; F:GO:0005524; F:GO:0008270; F:GO:0016743; C:GO:0009317; C:GO:0009570 | P:fatty acid biosynthetic process; P:malonyl-CoA biosynthetic process; F:acetyl-CoA carboxylase activity; F:ATP binding; F:zinc ion binding; F:carboxyl- or carbamoyltransferase activity; C:acetyl-CoA carboxylase complex; C:chloroplast stroma | Acetyl-CoA carboxylase; Transferring one-carbon groups | P:fatty acid biosynthetic process; F:acetyl-CoA carboxylase activity; C:acetyl-CoA carboxylase complex | view details | ||
| JN090102.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KY528596.1 | Acorus calamus | GTPase HflX | 1284 | F:GO:0003729; F:GO:0005525; F:GO:0043022; F:GO:0046872; C:GO:0005737 | F:mRNA binding; F:GTP binding; F:ribosome binding; F:metal ion binding; C:cytoplasm | F:GTP binding | view details | |||
| JF953048.1 | Acorus calamus | maturase K | 723 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| EU814664.1 | Acorus calamus | NADH dehydrogenase subunit F | 2011 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| LC219066.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| X84107.1 | Acorus calamus | ribosomal protein S4 | 588 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0009507; C:GO:0015935 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:chloroplast; C:small ribosomal subunit | P:translation; F:RNA binding; F:structural constituent of ribosome; F:rRNA binding; C:small ribosomal subunit | view details | |||
| HQ267432.1 | Acorus calamus | transport membrane protein | 579 | P:GO:0043953; P:GO:0065002; F:GO:0009977; C:GO:0005739; C:GO:0016021; C:GO:0033281 | P:protein transport by the Tat complex; P:intracellular protein transmembrane transport; F:proton motive force dependent protein transmembrane transporter activity; C:mitochondrion; C:integral component of membrane; C:TAT protein transport complex | Translocases | C:integral component of membrane | view details | ||
| KP083070.1 | Acorus calamus | phytochrome C | 1149 | P:GO:0006355; P:GO:0009584; P:GO:0018298; F:GO:0009881 | P:regulation of transcription, DNA-templated; P:detection of visible light; P:protein-chromophore linkage; F:photoreceptor activity | P:regulation of transcription, DNA-templated; P:detection of visible light; P:protein-chromophore linkage; F:protein binding | view details | |||
| KM399287.1 | Acorus calamus | carbohydrate-binding-like fold protein | 3120 | P:GO:0006950; F:GO:0003729; F:GO:0030246; C:GO:0016021 | P:response to stress; F:mRNA binding; F:carbohydrate binding; C:integral component of membrane | F:carbohydrate binding | view details | |||
| EU814658.1 | Acorus calamus | maturase K | 1043 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KJ667638.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 696 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY536842.1 | Acorus calamus | alpha/beta-Hydrolases superfamily protein | 858 | F:GO:0016787; C:GO:0016021 | F:hydrolase activity; C:integral component of membrane | Hydrolases | no GO terms | view details | ||
| MN481078.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY539214.1 | Acorus calamus | probable arabinosyltransferase ARAD1 | 930 | P:GO:0006486; F:GO:0016757; C:GO:0000139; C:GO:0016021 | P:protein glycosylation; F:glycosyltransferase activity; C:Golgi membrane; C:integral component of membrane | Glycosyltransferases | P:protein glycosylation; F:glycosyltransferase activity | view details | ||
| KM400318.1 | Acorus calamus | S-adenosyl-L-methionine-dependent methyltransferase superfamily protein | 993 | P:GO:0001510; F:GO:0003723; F:GO:0008168 | P:RNA methylation; F:RNA binding; F:methyltransferase activity | Transferring one-carbon groups | P:RNA methylation; F:methyltransferase activity | view details | ||
| KU127421.1 | Acorus calamus | maturase K | 808 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| MN481060.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO002062.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 150 | no GO terms | view details | |||||
| KM398368.1 | Acorus calamus | LOS4 | 1257 | F:GO:0003723; F:GO:0003724; F:GO:0005524; F:GO:0016787; C:GO:0009507 | F:RNA binding; F:RNA helicase activity; F:ATP binding; F:hydrolase activity; C:chloroplast | RNA helicase | F:nucleic acid binding; F:RNA helicase activity; F:ATP binding | view details | ||
| HO002067.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| JF953044.1 | Acorus calamus | maturase K | 738 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| FJ875013.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| KM399608.1 | Acorus calamus | Hus1-like protein | 936 | P:GO:0000077; C:GO:0005730; C:GO:0030896 | P:DNA damage checkpoint signaling; C:nucleolus; C:checkpoint clamp complex | P:DNA damage checkpoint signaling; C:nucleolus; C:checkpoint clamp complex | view details | |||
| KY539834.1 | Acorus calamus | DEAD-box ATP-dependent RNA helicase 1 | 1488 | F:GO:0003676; F:GO:0004386; F:GO:0005524; C:GO:0005634 | F:nucleic acid binding; F:helicase activity; F:ATP binding; C:nucleus | F:nucleic acid binding; F:ATP binding | view details | |||
| KY539448.1 | Acorus calamus | electron transfer flavoprotein subunit alpha, mitochondrial | 951 | P:GO:0022900; P:GO:0033539; F:GO:0009055; F:GO:0050660; C:GO:0005759 | P:electron transport chain; P:fatty acid beta-oxidation using acyl-CoA dehydrogenase; F:electron transfer activity; F:flavin adenine dinucleotide binding; C:mitochondrial matrix | Oxidoreductases | F:electron transfer activity; F:flavin adenine dinucleotide binding | view details | ||
| KY530892.1 | Acorus calamus | DNA replication complex GINS protein PSF1 | 276 | P:GO:1902983; C:GO:0000811 | P:DNA strand elongation involved in mitotic DNA replication; C:GINS complex | P:DNA replication; C:GINS complex | view details | |||
| KY527245.1 | Acorus calamus | pollen Ole e 1 allergen and extensin family protein | 363 | no GO terms | view details | |||||
| KY531471.1 | Acorus calamus | probable uridine nucleosidase 2 | 939 | P:GO:0006152; F:GO:0047724; C:GO:0005829 | P:purine nucleoside catabolic process; F:inosine nucleosidase activity; C:cytosol | Inosine nucleosidase; Purine nucleosidase | no GO terms | view details | ||
| KY541499.1 | Acorus calamus | NADH-cytochrome b5 reductase-like protein | 975 | F:GO:0004128; F:GO:0009703 | F:cytochrome-b5 reductase activity, acting on NAD(P)H; F:nitrate reductase (NADH) activity | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Cytochrome-b5 reductase; Nitrate reductase | F:oxidoreductase activity | view details | ||
| FJ875011.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM398206.1 | Acorus calamus | transducin/WD40 repeat-like superfamily protein | 1359 | C:GO:0005634; C:GO:0080008 | C:nucleus; C:Cul4-RING E3 ubiquitin ligase complex | F:protein binding | view details | |||
| KY524786.1 | Acorus calamus | nucleolar protein 6 isoform X2 | 2388 | P:GO:0006364; P:GO:0006409; C:GO:0032040; C:GO:0032545; C:GO:0034456 | P:rRNA processing; P:tRNA export from nucleus; C:small-subunit processome; C:CURI complex; C:UTP-C complex | no GO terms | view details | |||
| JF940682.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 615 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY537500.1 | Acorus calamus | protein ABCI7, chloroplastic | 495 | P:GO:0009793; P:GO:0010027; P:GO:0016226; C:GO:0009507; C:GO:0009536 | P:embryo development ending in seed dormancy; P:thylakoid membrane organization; P:iron-sulfur cluster assembly; C:chloroplast; C:plastid | P:iron-sulfur cluster assembly | view details | |||
| HO002039.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 236 | no GO terms | view details | |||||
| MN481058.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY541075.1 | Acorus calamus | Phosphoserine phosphatase | 813 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JF953046.1 | Acorus calamus | maturase K | 738 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KY528486.1 | Acorus calamus | protein Iojap-related, mitochondrial | 402 | P:GO:0017148; P:GO:0090071; F:GO:0043023 | P:negative regulation of translation; P:negative regulation of ribosome biogenesis; F:ribosomal large subunit binding | no GO terms | view details | |||
| DQ008884.1 | Acorus calamus | NADH dehydrogenase subunit F | 2032 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| KY541308.1 | Acorus calamus | DEAD-box ATP-dependent RNA helicase 13 isoform X1 | 1857 | F:GO:0003723; F:GO:0003724; F:GO:0005524; F:GO:0016787; C:GO:0005730 | F:RNA binding; F:RNA helicase activity; F:ATP binding; F:hydrolase activity; C:nucleolus | RNA helicase | F:nucleic acid binding; F:RNA helicase activity; F:ATP binding | view details | ||
| JN090103.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| EU814655.1 | Acorus calamus | maturase K | 1088 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KM397663.1 | Acorus calamus | pseudouridine synthase family protein | 930 | P:GO:0001522; P:GO:0006364; F:GO:0003723; F:GO:0009982 | P:pseudouridine synthesis; P:rRNA processing; F:RNA binding; F:pseudouridine synthase activity | Intramolecular transferases | P:pseudouridine synthesis; P:RNA modification; F:RNA binding; F:pseudouridine synthase activity | view details | ||
| KY531797.1 | Acorus calamus | E3 ubiquitin-protein ligase RNF14 | 1275 | P:GO:0000209; P:GO:0032436; F:GO:0031624; F:GO:0061630; C:GO:0000151; C:GO:0005737 | P:protein polyubiquitination; P:positive regulation of proteasomal ubiquitin-dependent protein catabolic process; F:ubiquitin conjugating enzyme binding; F:ubiquitin protein ligase activity; C:ubiquitin ligase complex; C:cytoplasm | Transferases | P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:protein binding | view details | ||
| LC461851.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JF953049.1 | Acorus calamus | maturase K | 729 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KY533771.1 | Acorus calamus | chitinase domain-containing protein 1 | 1218 | P:GO:0005975; P:GO:0006629; F:GO:0008061; F:GO:0008970; F:GO:0070492; C:GO:0005576; C:GO:0016021 | P:carbohydrate metabolic process; P:lipid metabolic process; F:chitin binding; F:phospholipase A1 activity; F:oligosaccharide binding; C:extracellular region; C:integral component of membrane | Phospholipase A(1); Carboxylesterase | P:carbohydrate metabolic process; F:chitin binding | view details | ||
| HO002047.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 293 | no GO terms | view details | |||||
| HQ267474.1 | Acorus calamus | NADH dehydrogenase subunit 5 | 1095 | P:GO:0042773; F:GO:0008137; C:GO:0005743; C:GO:0009536; C:GO:0009579; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; C:mitochondrial inner membrane; C:plastid; C:thylakoid; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| KY535656.1 | Acorus calamus | probable leucine-rich repeat receptor-like protein kinase At1g68400 | 1179 | P:GO:0016310; F:GO:0000166; F:GO:0016301; C:GO:0016020 | P:phosphorylation; F:nucleotide binding; F:kinase activity; C:membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| MN481070.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY526791.1 | Acorus calamus | vacuolar protein-sorting-associated protein 11 homolog isoform X1 | 2796 | P:GO:0006886; P:GO:0006904; P:GO:0007032; P:GO:0007033; P:GO:0048284; F:GO:0030674; F:GO:0046872; C:GO:0009705; C:GO:0030897; C:GO:0033263 | P:intracellular protein transport; P:vesicle docking involved in exocytosis; P:endosome organization; P:vacuole organization; P:organelle fusion; F:protein-macromolecule adaptor activity; F:metal ion binding; C:plant-type vacuole membrane; C:HOPS complex; C:CORVET complex | P:intracellular protein transport; P:vesicle-mediated transport; F:protein binding | view details | |||
| KM398747.1 | Acorus calamus | translation initiation factor 2 | 1929 | P:GO:0006413; F:GO:0003743; F:GO:0003924; F:GO:0005525; C:GO:0009507 | P:translational initiation; F:translation initiation factor activity; F:GTPase activity; F:GTP binding; C:chloroplast | Acting on acid anhydrides | P:translational initiation; F:translation initiation factor activity; F:GTPase activity; F:GTP binding | view details | ||
| KY537731.1 | Acorus calamus | PREDICTED: uncharacterized protein LOC104234818 isoform X2 | 735 | no GO terms | view details | |||||
| MN481069.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| FM882132.1 | Acorus calamus | WUSCHEL-related homeobox 11 | 161 | P:GO:0006355; P:GO:0010311; P:GO:0048830; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:lateral root formation; P:adventitious root development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:adventitious root development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| HB437553.1 | Acorus calamus | ribosomal protein S11 | 417 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0005840; C:GO:0009507 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:ribosome; C:chloroplast | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| KX344669.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 677 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM398262.1 | Acorus calamus | RNA ligase/cyclic nucleotide phosphodiesterase family protein | 471 | F:GO:0004112; F:GO:0016874 | F:cyclic-nucleotide phosphodiesterase activity; F:ligase activity | Acting on ester bonds; Ligases | F:cyclic-nucleotide phosphodiesterase activity | view details | ||
| KM398094.1 | Acorus calamus | nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein | 897 | P:GO:0006107; P:GO:0006108; P:GO:0006528; P:GO:0006541; F:GO:0016746; F:GO:0050152 | P:oxaloacetate metabolic process; P:malate metabolic process; P:asparagine metabolic process; P:glutamine metabolic process; F:acyltransferase activity; F:omega-amidase activity | Acyltransferases; Omega-amidase | P:nitrogen compound metabolic process; F:hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | view details | ||
| MN481072.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| M91625.2 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1324 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| GQ434259.1 | Acorus calamus | maturase K | 792 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JF940684.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 596 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY532363.1 | Acorus calamus | DEAD-box ATP-dependent RNA helicase 50 | 1749 | F:GO:0000166; F:GO:0003723; F:GO:0004386; C:GO:0005737; C:GO:0043229 | F:nucleotide binding; F:RNA binding; F:helicase activity; C:cytoplasm; C:intracellular organelle | F:nucleic acid binding; F:RNA helicase activity; F:ATP binding | view details | |||
| KY527224.1 | Acorus calamus | molybdenum cofactor sulfurase isoform X2 | 2463 | P:GO:0006777; P:GO:0019752; F:GO:0008265; F:GO:0008483; F:GO:0016829; F:GO:0030151; F:GO:0030170; F:GO:0102867 | P:Mo-molybdopterin cofactor biosynthetic process; P:carboxylic acid metabolic process; F:Mo-molybdopterin cofactor sulfurase activity; F:transaminase activity; F:lyase activity; F:molybdenum ion binding; F:pyridoxal phosphate binding; F:molybdenum cofactor sulfurtransferase activity | Transferring nitrogenous groups; Molybdenum cofactor sulfurtransferase; Lyases | P:Mo-molybdopterin cofactor biosynthetic process; F:catalytic activity; F:Mo-molybdopterin cofactor sulfurase activity; F:molybdenum ion binding; F:pyridoxal phosphate binding | view details | ||
| KY528799.1 | Acorus calamus | pentatricopeptide repeat-containing protein At4g31850, chloroplastic | 3297 | F:protein binding | view details | |||||
| KY531225.1 | Acorus calamus | general transcription and DNA repair factor IIH subunit TFB4 | 612 | P:GO:0006289; P:GO:0070816; F:GO:0046872; C:GO:0000439; C:GO:0005675 | P:nucleotide-excision repair; P:phosphorylation of RNA polymerase II C-terminal domain; F:metal ion binding; C:transcription factor TFIIH core complex; C:transcription factor TFIIH holo complex | P:nucleotide-excision repair; P:regulation of transcription, DNA-templated; C:transcription factor TFIIH core complex | view details | |||
| KM397439.1 | Acorus calamus | structural maintenance of chromosomes protein 1 | 3153 | P:GO:0007064; P:GO:0051321; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0008278; C:GO:0009507 | P:mitotic sister chromatid cohesion; P:meiotic cell cycle; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:cohesin complex; C:chloroplast | Acting on acid anhydrides | P:chromosome organization; F:protein binding; F:ATP binding; F:ATP hydrolysis activity; C:chromosome | view details | ||
| KY540222.1 | Acorus calamus | mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 | 1758 | P:GO:0005975; P:GO:0006491; P:GO:0030433; F:GO:0004571; F:GO:0005509; C:GO:0005783; C:GO:0016021 | P:carbohydrate metabolic process; P:N-glycan processing; P:ubiquitin-dependent ERAD pathway; F:mannosyl-oligosaccharide 1,2-alpha-mannosidase activity; F:calcium ion binding; C:endoplasmic reticulum; C:integral component of membrane | Mannosyl-oligosaccharide 1,2-alpha-mannosidase; Alpha-mannosidase | P:carbohydrate metabolic process; F:mannosyl-oligosaccharide 1,2-alpha-mannosidase activity; F:calcium ion binding; C:membrane | view details | ||
| KY534862.1 | Acorus calamus | putative hydrolase C777.06c | 858 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| KY540192.1 | Acorus calamus | sphingolipid delta(4)-desaturase DES1-like | 936 | P:GO:0046513; F:GO:0042284; C:GO:0005789; C:GO:0016021 | P:ceramide biosynthetic process; F:sphingolipid delta-4 desaturase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | Sphingolipid 4-desaturase | P:lipid metabolic process; P:sphingolipid biosynthetic process; F:sphingolipid delta-4 desaturase activity; C:integral component of membrane | view details | ||
| MN481066.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| AM889506.1 | Acorus calamus | acetyl-CoA carboxylase beta subunit | 381 | P:GO:0006633; P:GO:2001295; F:GO:0003989; F:GO:0005524; F:GO:0008270; F:GO:0016743; C:GO:0009317; C:GO:0009570 | P:fatty acid biosynthetic process; P:malonyl-CoA biosynthetic process; F:acetyl-CoA carboxylase activity; F:ATP binding; F:zinc ion binding; F:carboxyl- or carbamoyltransferase activity; C:acetyl-CoA carboxylase complex; C:chloroplast stroma | Acetyl-CoA carboxylase; Transferring one-carbon groups | P:fatty acid biosynthetic process; F:acetyl-CoA carboxylase activity; C:acetyl-CoA carboxylase complex | view details | ||
| KM399884.1 | Acorus calamus | protein arginine methyltransferase 6 | 990 | P:GO:0035246; F:GO:0016274 | P:peptidyl-arginine N-methylation; F:protein-arginine N-methyltransferase activity | Transferring one-carbon groups | P:peptidyl-arginine methylation; F:protein-arginine N-methyltransferase activity | view details | ||
| KY530346.1 | Acorus calamus | pentatricopeptide repeat-containing protein At1g79540 | 1776 | F:protein binding | view details | |||||
| HO002053.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 146 | no GO terms | view details | |||||
| KY527002.1 | Acorus calamus | peroxisome biogenesis protein 6 | 2166 | P:GO:0016558; F:GO:0016887; C:GO:0005778; C:GO:0005829 | P:protein import into peroxisome matrix; F:ATP hydrolysis activity; C:peroxisomal membrane; C:cytosol | Acting on acid anhydrides | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JN090021.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| AJ235381.2 | Acorus calamus | ATP synthase CF1 beta subunit | 1428 | P:GO:0015986; F:GO:0005524; F:GO:0046933; F:GO:0046961; C:GO:0009535; C:GO:0045261 | P:ATP synthesis coupled proton transport; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:chloroplast thylakoid membrane; C:proton-transporting ATP synthase complex, catalytic core F(1) | H(+)-exporting diphosphatase; H(+)-transporting two-sector ATPase; Ligases | no IPS match | view details | ||
| MN481073.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO002060.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY532662.1 | Acorus calamus | transmembrane protein 135 homolog | 1275 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| KY528380.1 | Acorus calamus | dihydrodipicolinate reductase-like protein CRR1, chloroplastic | 867 | P:GO:0009089; P:GO:0019877; F:GO:0008839; C:GO:0009570 | P:lysine biosynthetic process via diaminopimelate; P:diaminopimelate biosynthetic process; F:4-hydroxy-tetrahydrodipicolinate reductase; C:chloroplast stroma | 4-hydroxy-tetrahydrodipicolinate reductase; Acting on the CH-CH group of donors | P:lysine biosynthetic process via diaminopimelate; F:4-hydroxy-tetrahydrodipicolinate reductase | view details | ||
| KM398908.1 | Acorus calamus | iron-sulfur cluster biosynthesis family protein | 1335 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| MN481081.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| FM882139.1 | Acorus calamus | putative wuschel homeobox protein WOX13 | 140 | P:GO:0006355; F:GO:0000976; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:transcription cis-regulatory region binding; F:DNA-binding transcription factor activity; C:nucleus | F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| MH069738.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 605 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO002065.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| HO002071.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY527568.1 | Acorus calamus | AFG1-like ATPase | 450 | F:GO:0005524; F:GO:0016887; C:GO:0005739 | F:ATP binding; F:ATP hydrolysis activity; C:mitochondrion | Acting on acid anhydrides | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| KF613092.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO002051.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 293 | no GO terms | view details | |||||
| KX344528.1 | Acorus calamus | maturase K | 696 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KY529490.1 | Acorus calamus | nucleolar complex protein 3 homolog | 1986 | C:GO:0005634; C:GO:0005730 | C:nucleus; C:nucleolus | no GO terms | view details | |||
| MN481065.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY529798.1 | Acorus calamus | elongation factor G-2, chloroplastic | 2196 | P:GO:0006414; P:GO:0032543; P:GO:0032790; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0005739; C:GO:0009507 | P:translational elongation; P:mitochondrial translation; P:ribosome disassembly; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:mitochondrion; C:chloroplast | Acting on acid anhydrides | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:chloroplast | view details | ||
| KY530091.1 | Acorus calamus | mitochondrial carrier protein CoAc2 | 963 | P:GO:1990559; F:GO:0015228; C:GO:0005743; C:GO:0016021 | P:mitochondrial coenzyme A transmembrane transport; F:coenzyme A transmembrane transporter activity; C:mitochondrial inner membrane; C:integral component of membrane | Translocases | P:transmembrane transport | view details | ||
| MG228099.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM400157.1 | Acorus calamus | AT1G65230-like protein | 687 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| HO002048.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 293 | no GO terms | view details | |||||
| KY524670.1 | Acorus calamus | soluble starch synthase 1, chloroplastic/amyloplastic isoform X1 | 999 | P:GO:0009960; P:GO:0010021; P:GO:0019252; F:GO:0004373; F:GO:0009011; C:GO:0009501; C:GO:0009507 | P:endosperm development; P:amylopectin biosynthetic process; P:starch biosynthetic process; F:glycogen (starch) synthase activity; F:starch synthase activity; C:amyloplast; C:chloroplast | Starch synthase; Glycogen(starch) synthase | F:glycogen (starch) synthase activity; F:glycosyltransferase activity | view details | ||
| KY531261.1 | Acorus calamus | dehydrogenase/reductase SDR family member 12 | 942 | F:GO:0016491; C:GO:0016021 | F:oxidoreductase activity; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity | view details | ||
| KY538896.1 | Acorus calamus | protein ecdysoneless homolog | 1428 | C:GO:0005634 | C:nucleus | no GO terms | view details | |||
| KY541534.1 | Acorus calamus | dehydrogenase/reductase SDR family member 12 | 942 | F:GO:0016491; C:GO:0016021 | F:oxidoreductase activity; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity | view details | ||
| KM399991.1 | Acorus calamus | ATP synthase protein I-related protein | 885 | P:GO:0033614; C:GO:0009535; C:GO:0016021 | P:chloroplast proton-transporting ATP synthase complex assembly; C:chloroplast thylakoid membrane; C:integral component of membrane | no GO terms | view details | |||
| KY536704.1 | Acorus calamus | origin of replication complex subunit 6 | 456 | P:GO:0006270; P:GO:0008610; P:GO:0009555; F:GO:0003677; F:GO:0005506; F:GO:0016491; C:GO:0005664; C:GO:0016020 | P:DNA replication initiation; P:lipid biosynthetic process; P:pollen development; F:DNA binding; F:iron ion binding; F:oxidoreductase activity; C:nuclear origin of replication recognition complex; C:membrane | Oxidoreductases | P:DNA replication; C:nuclear origin of replication recognition complex | view details | ||
| KY530685.1 | Acorus calamus | protein root UVB sensitive 3 | 450 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| KY529917.1 | Acorus calamus | probable histone acetyltransferase type B catalytic subunit | 1113 | P:GO:0016573; F:GO:0004402; F:GO:0016830 | P:histone acetylation; F:histone acetyltransferase activity; F:carbon-carbon lyase activity | Carbon-carbon lyases; Histone acetyltransferase | P:chromatin organization; P:histone acetylation; P:subtelomeric heterochromatin assembly; F:histone acetyltransferase activity; F:N-acetyltransferase activity; C:nucleus | view details | ||
| KX347061.2 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 573 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY527950.1 | Acorus calamus | probable inactive shikimate kinase like 2, chloroplastic | 723 | P:GO:0016310; F:GO:0016301; F:GO:0016787; C:GO:0009507 | P:phosphorylation; F:kinase activity; F:hydrolase activity; C:chloroplast | Transferring phosphorus-containing groups; Hydrolases | no GO terms | view details | ||
| KM399072.1 | Acorus calamus | RNA polymerase II transcription mediator | 1542 | P:GO:0006357; P:GO:0035196; F:GO:0003712; C:GO:0016592 | P:regulation of transcription by RNA polymerase II; P:production of miRNAs involved in gene silencing by miRNA; F:transcription coregulator activity; C:mediator complex | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | view details | |||
| HO002056.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| HO002042.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 294 | no GO terms | view details | |||||
| KY540973.1 | Acorus calamus | ribonuclease E/G-like protein, chloroplastic isoform X1 | 2823 | P:GO:0006364; P:GO:0090501; F:GO:0004540; F:GO:0030246; C:GO:0005737 | P:rRNA processing; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity; F:carbohydrate binding; C:cytoplasm | Acting on ester bonds | P:RNA processing; F:RNA binding; F:ribonuclease activity; F:carbohydrate binding; F:starch binding | view details | ||
| KM399019.1 | Acorus calamus | protein NRT1/ PTR FAMILY 6.1 | 1692 | P:GO:0006857; P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:oligopeptide transport; P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| KM399663.1 | Acorus calamus | ABC1-like kinase | 1848 | P:GO:0006468; P:GO:0006995; P:GO:0009414; P:GO:0009644; P:GO:0010114; P:GO:0050821; P:GO:0080177; P:GO:0080183; P:GO:1902171; F:GO:0004672; F:GO:0005524; C:GO:0010287 | P:protein phosphorylation; P:cellular response to nitrogen starvation; P:response to water deprivation; P:response to high light intensity; P:response to red light; P:protein stabilization; P:plastoglobule organization; P:response to photooxidative stress; P:regulation of tocopherol cyclase activity; F:protein kinase activity; F:ATP binding; C:plastoglobule | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| AF123799.1 | Acorus calamus | NADH dehydrogenase subunit F | 1288 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| JN090019.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY527433.1 | Acorus calamus | serine/threonine-protein kinase ATR | 1872 | P:GO:0000077; P:GO:0000723; P:GO:0006281; P:GO:0006468; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310; C:GO:0005634 | P:DNA damage checkpoint signaling; P:telomere maintenance; P:DNA repair; P:protein phosphorylation; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity; C:nucleus | Dual-specificity kinase | F:protein binding; F:kinase activity | view details | ||
| HO002066.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| EU814663.1 | Acorus calamus | NADH dehydrogenase subunit F | 1922 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| HO002058.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 148 | no GO terms | view details | |||||
| KM400046.1 | Acorus calamus | EMB1873 protein | 1062 | P:GO:0009446; F:GO:0004668; F:GO:0047632 | P:putrescine biosynthetic process; F:protein-arginine deiminase activity; F:agmatine deiminase activity | Agmatine deiminase; Protein-arginine deiminase | P:putrescine biosynthetic process; F:protein-arginine deiminase activity; F:agmatine deiminase activity | view details | ||
| KY535772.1 | Acorus calamus | bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial | 1755 | P:GO:0009102; F:GO:0000166; F:GO:0004015; F:GO:0004141; F:GO:0043168; C:GO:0005739 | P:biotin biosynthetic process; F:nucleotide binding; F:adenosylmethionine-8-amino-7-oxononanoate transaminase activity; F:dethiobiotin synthase activity; F:anion binding; C:mitochondrion | Dethiobiotin synthase; Adenosylmethionine--8-amino-7-oxononanoate transaminase | F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| KY537161.1 | Acorus calamus | probable inactive shikimate kinase like 2, chloroplastic | 723 | P:GO:0016310; F:GO:0016301; F:GO:0016787; C:GO:0009507 | P:phosphorylation; F:kinase activity; F:hydrolase activity; C:chloroplast | Transferring phosphorus-containing groups; Hydrolases | no GO terms | view details | ||
| KY537951.1 | Acorus calamus | Cysteine desulfurase, mitochondrial | 1368 | P:GO:0044571; P:GO:1901566; F:GO:0030170; F:GO:0031071; F:GO:0046872; F:GO:0051536; C:GO:0005739; C:GO:0005829 | P:[2Fe-2S] cluster assembly; P:organonitrogen compound biosynthetic process; F:pyridoxal phosphate binding; F:cysteine desulfurase activity; F:metal ion binding; F:iron-sulfur cluster binding; C:mitochondrion; C:cytosol | Cysteine desulfurase | P:[2Fe-2S] cluster assembly; F:catalytic activity; F:pyridoxal phosphate binding; F:cysteine desulfurase activity | view details | ||
| HO002057.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KM397716.1 | Acorus calamus | pollen Ole e 1 allergen and extensin family protein | 360 | no GO terms | view details | |||||
| KY528617.1 | Acorus calamus | exosome complex exonuclease RRP44 homolog A | 2661 | P:GO:0016075; P:GO:0090503; F:GO:0000175; F:GO:0003723; F:GO:0004519; C:GO:0000176; C:GO:0000177 | P:rRNA catabolic process; P:RNA phosphodiester bond hydrolysis, exonucleolytic; F:3 -5-exoribonuclease activity; F:RNA binding; F:endonuclease activity; C:nuclear exosome (RNase complex); C:cytoplasmic exosome (RNase complex) | Acting on ester bonds; Acting on ester bonds | F:RNA binding; F:ribonuclease activity | view details | ||
| X59457.1 | Acorus calamus | hypothetical protein RhiirA1_354704 | 254 | no IPS match | view details | |||||
| JN090098.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KF632754.1 | Acorus calamus | cytochrome c biogenesis B | 579 | P:GO:0006412; P:GO:0015886; P:GO:0017004; F:GO:0003735; F:GO:0015232; C:GO:0005634; C:GO:0005739; C:GO:0005840; C:GO:0043190 | P:translation; P:heme transport; P:cytochrome complex assembly; F:structural constituent of ribosome; F:heme transmembrane transporter activity; C:nucleus; C:mitochondrion; C:ribosome; C:ATP-binding cassette (ABC) transporter complex | Translocases | P:heme transport; P:cytochrome complex assembly; F:heme transmembrane transporter activity; C:membrane | view details | ||
| HO002041.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 269 | no GO terms | view details | |||||
| EU814669.1 | Acorus calamus | NADH dehydrogenase subunit F | 1998 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| KM398690.1 | Acorus calamus | galactose mutarotase-like superfamily protein | 642 | P:GO:0005975; F:GO:0030246; F:GO:0047938; C:GO:0005737 | P:carbohydrate metabolic process; F:carbohydrate binding; F:glucose-6-phosphate 1-epimerase activity; C:cytoplasm | Glucose-6-phosphate 1-epimerase | P:carbohydrate metabolic process; F:catalytic activity; F:isomerase activity; F:carbohydrate binding | view details | ||
| KY525668.1 | Acorus calamus | Muniscin C-terminal mu homology domain | 1725 | P:GO:0009555; P:GO:0072583 | P:pollen development; P:clathrin-dependent endocytosis | no GO terms | view details | |||
| LC219310.1 | Acorus calamus | maturase K | 786 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| GQ435780.1 | Acorus calamus | cytochrome c heme attachment protein | 376 | P:GO:0017004; F:GO:0020037; C:GO:0009535; C:GO:0016021 | P:cytochrome complex assembly; F:heme binding; C:chloroplast thylakoid membrane; C:integral component of membrane | P:cytochrome complex assembly; F:heme binding | view details | |||
| KX347060.2 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 573 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY534742.1 | Acorus calamus | DEAD-box ATP-dependent RNA helicase 5 | 1305 | P:GO:0006364; F:GO:0003723; F:GO:0003724; F:GO:0005524; F:GO:0016787; C:GO:0005730; C:GO:0009507 | P:rRNA processing; F:RNA binding; F:RNA helicase activity; F:ATP binding; F:hydrolase activity; C:nucleolus; C:chloroplast | RNA helicase | F:nucleic acid binding; F:RNA helicase activity; F:ATP binding | view details | ||
| KY526827.1 | Acorus calamus | Calmodulin-interacting protein 111 | 690 | F:GO:0005524; F:GO:0016887 | F:ATP binding; F:ATP hydrolysis activity | Acting on acid anhydrides | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| KM398423.1 | Acorus calamus | fatty acid amide hydrolase | 1548 | P:GO:0042742; P:GO:0070291; F:GO:0047412; C:GO:0005886 | P:defense response to bacterium; P:N-acylethanolamine metabolic process; F:N-(long-chain-acyl)ethanolamine deacylase activity; C:plasma membrane | N-(long-chain-acyl)ethanolamine deacylase | F:catalytic activity | view details | ||
| KY539197.1 | Acorus calamus | sialyltransferase-like protein 2 | 1317 | P:GO:0006486; P:GO:0009846; P:GO:0009860; P:GO:0097503; F:GO:0008373; C:GO:0000139; C:GO:0016021 | P:protein glycosylation; P:pollen germination; P:pollen tube growth; P:sialylation; F:sialyltransferase activity; C:Golgi membrane; C:integral component of membrane | Glycosyltransferases | P:protein glycosylation; F:sialyltransferase activity | view details | ||
| KM399718.1 | Acorus calamus | eyes absent-like protein | 738 | P:GO:0035335; F:GO:0030946; F:GO:0046872 | P:peptidyl-tyrosine dephosphorylation; F:protein tyrosine phosphatase activity, metal-dependent; F:metal ion binding | Protein-tyrosine-phosphatase | P:multicellular organism development; F:protein tyrosine phosphatase activity | view details | ||
| HO002055.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| MN481079.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| FM882138.1 | Acorus calamus | wuschel | 164 | P:GO:0006355; P:GO:0099402; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| KY535564.1 | Acorus calamus | probable bifunctional TENA-E protein | 645 | P:GO:0006772; C:GO:0005829 | P:thiamine metabolic process; C:cytosol | no GO terms | view details | |||
| KY525701.1 | Acorus calamus | cactin isoform X2 | 1671 | P:GO:0045292; F:GO:0003723; C:GO:0005681; C:GO:0005737 | P:mRNA cis splicing, via spliceosome; F:RNA binding; C:spliceosomal complex; C:cytoplasm | F:protein binding | view details | |||
| KY541165.1 | Acorus calamus | GTP-binding protein BRASSINAZOLE INSENSITIVE PALE GREEN 2, chloroplastic | 1539 | P:GO:0009742; P:GO:1901259; F:GO:0005525; F:GO:0016491; C:GO:0005739; C:GO:0009570 | P:brassinosteroid mediated signaling pathway; P:chloroplast rRNA processing; F:GTP binding; F:oxidoreductase activity; C:mitochondrion; C:chloroplast stroma | Oxidoreductases | F:GTP binding | view details | ||
| KX344595.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 642 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JN090100.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JF953045.1 | Acorus calamus | maturase K | 738 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KY535242.1 | Acorus calamus | pentatricopeptide repeat-containing protein At4g01030, mitochondrial | 1440 | F:GO:0008270 | F:zinc ion binding | F:protein binding; F:zinc ion binding | view details | |||
| MN481076.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY529346.1 | Acorus calamus | uncharacterized protein LOC109839589 | 360 | no GO terms | view details | |||||
| KM398635.1 | Acorus calamus | AT2G18410-like protein | 900 | P:GO:0002098; C:GO:0005634; C:GO:0005737; C:GO:0033588 | P:tRNA wobble uridine modification; C:nucleus; C:cytoplasm; C:elongator holoenzyme complex | P:tRNA wobble uridine modification; C:elongator holoenzyme complex | view details | |||
| KM397771.1 | Acorus calamus | homoserine kinase | 948 | P:GO:0009088; P:GO:0016310; F:GO:0004413; F:GO:0005524 | P:threonine biosynthetic process; P:phosphorylation; F:homoserine kinase activity; F:ATP binding | Homoserine kinase | P:threonine metabolic process; F:homoserine kinase activity; F:ATP binding | view details | ||
| KM400427.1 | Acorus calamus | AT4G29520-like protein | 504 | no GO terms | view details | |||||
| MN481063.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JN090101.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| FJ875016.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO002073.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY532022.1 | Acorus calamus | putative 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial | 2124 | P:GO:0005975; P:GO:0016310; F:GO:0005524; F:GO:0008270; F:GO:0016301; F:GO:0016491; F:GO:0016832; F:GO:0050661; F:GO:0051287 | P:carbohydrate metabolic process; P:phosphorylation; F:ATP binding; F:zinc ion binding; F:kinase activity; F:oxidoreductase activity; F:aldehyde-lyase activity; F:NADP binding; F:NAD binding | Carbon-carbon lyases; Oxidoreductases; Transferring phosphorus-containing groups | P:carbohydrate metabolic process; F:catalytic activity; F:zinc ion binding; F:aldehyde-lyase activity | view details | ||
| KX344596.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 563 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MN481080.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY532882.1 | Acorus calamus | YLP motif-containing protein 1 | 1362 | P:GO:0032204; C:GO:0005634 | P:regulation of telomere maintenance; C:nucleus | C:nucleus | view details | |||
| EU814665.1 | Acorus calamus | NADH dehydrogenase subunit F | 1925 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| AB040154.1 | Acorus calamus | maturase K | 1551 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JN090020.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY533574.1 | Acorus calamus | prolyl-tRNA synthetase associated domain-containing protein 1 | 918 | P:GO:0106074; F:GO:0002161; F:GO:0004812 | P:aminoacyl-tRNA metabolism involved in translational fidelity; F:aminoacyl-tRNA editing activity; F:aminoacyl-tRNA ligase activity | Forming carbon-oxygen bonds; Carboxylesterase | F:aminoacyl-tRNA editing activity | view details | ||
| KX346958.2 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 573 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY540639.1 | Acorus calamus | protein ROOT INITIATION DEFECTIVE 3-like | 1239 | F:protein binding | view details | |||||
| JN090018.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY540863.1 | Acorus calamus | molybdate-anion transporter isoform X1 | 1113 | P:GO:0015698; C:GO:0016020 | P:inorganic anion transport; C:membrane | P:molybdate ion transport; F:molybdate ion transmembrane transporter activity; C:integral component of membrane | view details | |||
| KY539961.1 | Acorus calamus | pentatricopeptide repeat-containing protein At5g39980, chloroplastic | 1587 | P:GO:0009658; P:GO:0031425; F:GO:0003729; C:GO:0009570; C:GO:0009941 | P:chloroplast organization; P:chloroplast RNA processing; F:mRNA binding; C:chloroplast stroma; C:chloroplast envelope | F:protein binding | view details | |||
| MN481082.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| FM882134.1 | Acorus calamus | wuschel related homeobox 2 | 161 | P:GO:0006355; P:GO:0099402; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| KY529076.1 | Acorus calamus | DNA repair protein RAD51 homolog 3 isoform X3 | 513 | P:GO:0000707; P:GO:0007131; P:GO:0007140; P:GO:0007143; P:GO:0016444; F:GO:0000400; F:GO:0005524; F:GO:0008094; F:GO:0008821; C:GO:0005657; C:GO:0033063; C:GO:0033065 | P:meiotic DNA recombinase assembly; P:reciprocal meiotic recombination; P:male meiotic nuclear division; P:female meiotic nuclear division; P:somatic cell DNA recombination; F:four-way junction DNA binding; F:ATP binding; F:ATP-dependent activity, acting on DNA; F:crossover junction endodeoxyribonuclease activity; C:replication fork; C:Rad51B-Rad51C-Rad51D-XRCC2 complex; C:Rad51C-XRCC3 complex | Acting on ester bonds; Crossover junction endodeoxyribonuclease | P:DNA repair; F:DNA binding; F:ATP binding; F:ATP-dependent activity, acting on DNA | view details | ||
| DQ008885.1 | Acorus calamus | NADH dehydrogenase subunit F | 2032 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| KF632783.1 | Acorus calamus | maturase K | 853 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KY535368.1 | Acorus calamus | protein NRT1/ PTR FAMILY 6.1 | 1764 | P:GO:0006857; P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:oligopeptide transport; P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| KY532138.1 | Acorus calamus | protein fluG isoform X1 | 1635 | P:GO:0006542; F:GO:0004356; F:GO:0016787 | P:glutamine biosynthetic process; F:glutamate-ammonia ligase activity; F:hydrolase activity | Hydrolases; Glutamine synthetase | P:glutamine biosynthetic process; P:nitrogen compound metabolic process; F:catalytic activity; F:glutamate-ammonia ligase activity; F:hydrolase activity | view details | ||
| FJ875012.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| GQ248071.1 | Acorus calamus | maturase K | 792 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| AB017314.1 | Acorus calamus | maturase K | 1109 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KY536496.1 | Acorus calamus | E3 ubiquitin-protein ligase SP1 | 1017 | P:GO:0006996; P:GO:0016567; F:GO:0004842; F:GO:0016874; F:GO:0046872; C:GO:0016021 | P:organelle organization; P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:ligase activity; F:metal ion binding; C:integral component of membrane | Transferases; Ligases | P:organelle organization; P:protein ubiquitination; F:ubiquitin-protein transferase activity | view details | ||
| KY529364.1 | Acorus calamus | electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial isoform X1 | 1602 | P:GO:0006552; P:GO:0009646; P:GO:0022904; F:GO:0004174; F:GO:0046872; F:GO:0051539; C:GO:0031305 | P:leucine catabolic process; P:response to absence of light; P:respiratory electron transport chain; F:electron-transferring-flavoprotein dehydrogenase activity; F:metal ion binding; F:4 iron, 4 sulfur cluster binding; C:integral component of mitochondrial inner membrane | Electron-transferring-flavoprotein dehydrogenase | P:electron transport chain; F:electron-transferring-flavoprotein dehydrogenase activity | view details | ||
| KY530462.1 | Acorus calamus | S-adenosyl-L-methionine-dependent methyltransferase superfamily protein | 1125 | P:GO:0001510; F:GO:0003723; F:GO:0008168 | P:RNA methylation; F:RNA binding; F:methyltransferase activity | Transferring one-carbon groups | P:RNA methylation; F:methyltransferase activity | view details | ||
| MN481067.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY532997.1 | Acorus calamus | F-box protein At5g52880 | 243 | C:GO:0005737 | C:cytoplasm | no GO terms | view details | |||
| KY528061.1 | Acorus calamus | 2-(3-amino-3-carboxypropyl)histidine synthase subunit 2 | 1539 | P:GO:0017183; F:GO:0016740 | P:peptidyl-diphthamide biosynthetic process from peptidyl-histidine; F:transferase activity | Transferases | P:peptidyl-diphthamide biosynthetic process from peptidyl-histidine | view details | ||
| KY534584.1 | Acorus calamus | protein root UVB sensitive 1, chloroplastic | 1089 | P:GO:0010224; P:GO:0032502 | P:response to UV-B; P:developmental process | no GO terms | view details | |||
| KY534400.1 | Acorus calamus | eukaryotic translation initiation factor 2A | 1527 | P:GO:0006413; P:GO:0006417; F:GO:0000049; F:GO:0003729; F:GO:0003743; F:GO:0043022; C:GO:0022627 | P:translational initiation; P:regulation of translation; F:tRNA binding; F:mRNA binding; F:translation initiation factor activity; F:ribosome binding; C:cytosolic small ribosomal subunit | P:translational initiation; F:translation initiation factor activity; F:protein binding | view details | |||
| EU814657.1 | Acorus calamus | maturase K | 1118 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| JN090104.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| AJ344261.1 | Acorus calamus | photosystem I P700 apoprotein A1 | 1986 | P:GO:0015979; P:GO:0018298; P:GO:0022900; F:GO:0000287; F:GO:0009055; F:GO:0016168; F:GO:0051539; C:GO:0009522; C:GO:0009535; C:GO:0016021 | P:photosynthesis; P:protein-chromophore linkage; P:electron transport chain; F:magnesium ion binding; F:electron transfer activity; F:chlorophyll binding; F:4 iron, 4 sulfur cluster binding; C:photosystem I; C:chloroplast thylakoid membrane; C:integral component of membrane | Oxidoreductases | P:photosynthesis; F:metal ion binding; C:thylakoid; C:integral component of membrane | view details | ||
| KY536725.1 | Acorus calamus | protoheme IX farnesyltransferase, mitochondrial | 849 | P:GO:0045333; P:GO:0048034; F:GO:0008495; C:GO:0005739; C:GO:0016021 | P:cellular respiration; P:heme O biosynthetic process; F:protoheme IX farnesyltransferase activity; C:mitochondrion; C:integral component of membrane | Geranylgeranyl diphosphate synthase; Heme o synthase | P:heme biosynthetic process; F:protoheme IX farnesyltransferase activity; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:integral component of membrane | view details | ||
| HO002068.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 150 | no GO terms | view details | |||||
| LC387636.1 | Acorus calamus | trans-resveratrol di-O-methyltransferase | 1032 | P:GO:0032259; F:GO:0008171; F:GO:0046983 | P:methylation; F:O-methyltransferase activity; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| KY536380.1 | Acorus calamus | pentatricopeptide repeat-containing protein At1g09900 | 1686 | F:protein binding | view details | |||||
| KY535347.1 | Acorus calamus | histone-lysine N-methyltransferase ASHR1 isoform X1 | 1440 | P:GO:0005975; P:GO:0034968; F:GO:0016773; F:GO:0018024; F:GO:0046872; C:GO:0005634 | P:carbohydrate metabolic process; P:histone lysine methylation; F:phosphotransferase activity, alcohol group as acceptor; F:histone-lysine N-methyltransferase activity; F:metal ion binding; C:nucleus | Transferring phosphorus-containing groups; [Histone H3]-lysine(4) N-trimethyltransferase | F:protein binding | view details | ||
| KY525213.1 | Acorus calamus | pentatricopeptide repeat-containing protein At5g16420, mitochondrial | 981 | F:protein binding | view details | |||||
| HO002054.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KM399127.1 | Acorus calamus | uncoupling protein 3 | 813 | P:GO:0009409; P:GO:0055085; C:GO:0016021 | P:response to cold; P:transmembrane transport; C:integral component of membrane | P:transmembrane transport | view details | |||
| KM823798.1 | Acorus calamus | rhomboid-like protein 15 isoform X1 | 1311 | P:GO:0006508; F:GO:0004252; C:GO:0016021 | P:proteolysis; F:serine-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | F:serine-type endopeptidase activity; F:protein binding; C:integral component of membrane | view details | ||
| KM399503.1 | Acorus calamus | aspartyl protease family protein | 1227 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JN090017.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| DQ008869.1 | Acorus calamus | maturase K | 992 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KY538163.1 | Acorus calamus | protein DGS1, mitochondrial isoform X2 | 615 | P:GO:0019375; P:GO:0050665; C:GO:0005741; C:GO:0016021; C:GO:0032991 | P:galactolipid biosynthetic process; P:hydrogen peroxide biosynthetic process; C:mitochondrial outer membrane; C:integral component of membrane; C:protein-containing complex | no GO terms | view details | |||
| KY528909.1 | Acorus calamus | Protein sym-1 like | 972 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| HO002072.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY526129.1 | Acorus calamus | 39S ribosomal protein L45, mitochondrial isoform X1 | 750 | C:GO:0005739; C:GO:0005840 | C:mitochondrion; C:ribosome | no GO terms | view details | |||
| KM397986.1 | Acorus calamus | RabGAP/TBC domain-containing protein | 1650 | C:GO:0005737; C:GO:0016021 | C:cytoplasm; C:integral component of membrane | no GO terms | view details | |||
| KX346959.2 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 573 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY535986.1 | Acorus calamus | tubby-like protein 8 | 600 | no GO terms | view details | |||||
| GQ248538.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 553 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY530702.1 | Acorus calamus | RNA polymerase sigma factor sigC | 1281 | P:GO:2000142; F:GO:0003677; F:GO:0016987 | P:regulation of DNA-templated transcription, initiation; F:DNA binding; F:sigma factor activity | P:DNA-templated transcription, initiation; P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity | view details | |||
| MN481061.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX365297.1 | Acorus calamus | RNA polymerase C | 360 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| JN090016.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| Z80856.1 | Acorus calamus | NADH dehydrogenase subunit 2 | 444 | P:GO:0019684; P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0005886; C:GO:0009535; C:GO:0016021 | P:photosynthesis, light reaction; P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:plasma membrane; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | no GO terms | view details | ||
| KM400482.1 | Acorus calamus | embryo yellow protein | 2481 | P:GO:0006886; P:GO:0006890; P:GO:0007030; C:GO:0000139; C:GO:0017119 | P:intracellular protein transport; P:retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum; P:Golgi organization; C:Golgi membrane; C:Golgi transport complex | P:intracellular protein transport; C:Golgi transport complex | view details | |||
| KF613093.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY527800.1 | Acorus calamus | histone-lysine N-methyltransferase setd3 | 1269 | P:GO:0018026; F:GO:0016279 | P:peptidyl-lysine monomethylation; F:protein-lysine N-methyltransferase activity | Transferring one-carbon groups | F:protein binding | view details | ||
| AY007647.2 | Acorus calamus | NADH dehydrogenase subunit F | 2078 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| MN481059.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| LC219091.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY525330.1 | Acorus calamus | Structural maintenance of chromosomes protein 5 | 2811 | P:GO:0000724; P:GO:0007062; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0030915 | P:double-strand break repair via homologous recombination; P:sister chromatid cohesion; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:Smc5-Smc6 complex | Acting on acid anhydrides | P:double-strand break repair via homologous recombination; P:DNA repair; F:ATP binding; F:ATP hydrolysis activity; C:Smc5-Smc6 complex | view details | ||
| KM397931.1 | Acorus calamus | quasimodo 3 | 1515 | P:GO:0032259; P:GO:0042546; P:GO:0052546; F:GO:0008757; C:GO:0000139; C:GO:0016021 | P:methylation; P:cell wall biogenesis; P:cell wall pectin metabolic process; F:S-adenosylmethionine-dependent methyltransferase activity; C:Golgi membrane; C:integral component of membrane | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| DQ406951.1 | Acorus calamus | NADH dehydrogenase subunit 5 | 1076 | P:GO:0042773; F:GO:0008137; C:GO:0005739; C:GO:0009536; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; C:mitochondrion; C:plastid; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details | ||
| MN481064.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| AM889752.1 | Acorus calamus | NADH plastoquinone oxidoreductase subunit K | 216 | P:GO:0022900; F:GO:0008137; F:GO:0046872; F:GO:0048038; F:GO:0051539; C:GO:0009507; C:GO:0016021 | P:electron transport chain; F:NADH dehydrogenase (ubiquinone) activity; F:metal ion binding; F:quinone binding; F:4 iron, 4 sulfur cluster binding; C:chloroplast; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | F:iron-sulfur cluster binding | view details | ||
| JF940683.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 599 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY533332.1 | Acorus calamus | pentatricopeptide repeat-containing protein PNM1, mitochondrial | 1155 | C:GO:0043231 | C:intracellular membrane-bounded organelle | F:protein binding | view details | |||
| FM882135.1 | Acorus calamus | WUSCHEL-related homeobox 3B-like | 161 | P:GO:0006355; P:GO:0099402; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:plant organ development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| FM882133.1 | Acorus calamus | WUSCHEL-related homeobox 11 | 161 | P:GO:0006355; P:GO:0010311; P:GO:0048830; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:lateral root formation; P:adventitious root development; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:adventitious root development; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| KY538216.1 | Acorus calamus | uroporphyrinogen decarboxylase | 1080 | P:GO:0006782; P:GO:0015995; F:GO:0004853; C:GO:0009507 | P:protoporphyrinogen IX biosynthetic process; P:chlorophyll biosynthetic process; F:uroporphyrinogen decarboxylase activity; C:chloroplast | Uroporphyrinogen decarboxylase | P:porphyrin-containing compound biosynthetic process; F:uroporphyrinogen decarboxylase activity | view details | ||
| KY528280.1 | Acorus calamus | afadin- and alpha-actinin-binding protein isoform X1 | 1104 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| HO002061.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| KY530267.1 | Acorus calamus | cysteine synthase 2 | 1125 | P:GO:0006535; C:GO:0005737; C:GO:0016021 | P:cysteine biosynthetic process from serine; C:cytoplasm; C:integral component of membrane | no GO terms | view details | |||
| KM397383.1 | Acorus calamus | structural maintenance of chromosomes protein 2 | 2670 | P:GO:0000819; P:GO:0030261; P:GO:0051321; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0005694; C:GO:0009507 | P:sister chromatid segregation; P:chromosome condensation; P:meiotic cell cycle; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:chromosome; C:chloroplast | Acting on acid anhydrides | P:chromosome organization; F:protein binding; F:ATP binding; C:chromosome | view details | ||
| KY538055.1 | Acorus calamus | transcription termination factor MTERF4, chloroplastic | 1314 | P:GO:0006353; P:GO:0006355; P:GO:0032502; F:GO:0003690 | P:DNA-templated transcription, termination; P:regulation of transcription, DNA-templated; P:developmental process; F:double-stranded DNA binding | P:regulation of transcription, DNA-templated; F:double-stranded DNA binding | view details | |||
| JN090022.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 501 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY535793.1 | Acorus calamus | Lactamase B domain-containing protein | 1188 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| MN481077.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY533838.1 | Acorus calamus | DEAD-box ATP-dependent RNA helicase 22 | 1263 | F:GO:0000166; F:GO:0003723; F:GO:0003724 | F:nucleotide binding; F:RNA binding; F:RNA helicase activity | RNA helicase | F:nucleic acid binding; F:ATP binding | view details | ||
| LC461850.1 | Acorus calamus | maturase K | 732 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| DQ008868.1 | Acorus calamus | maturase K | 994 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KY526234.1 | Acorus calamus | WD40 repeat | 1173 | C:GO:0110165 | C:cellular anatomical entity | F:protein binding | view details | |||
| KY531016.1 | Acorus calamus | protein MCM10 homolog | 657 | P:GO:0006270; F:GO:0003688; F:GO:0003697; C:GO:0031298 | P:DNA replication initiation; F:DNA replication origin binding; F:single-stranded DNA binding; C:replication fork protection complex | P:DNA replication; P:DNA replication initiation; F:double-stranded DNA binding; F:single-stranded DNA binding; C:nucleus | view details | |||
| LC461849.1 | Acorus calamus | senescence-associated protein, putative | 339 | no IPS match | view details | |||||
| DQ008863.1 | Acorus calamus | maturase K | 969 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| KY536605.1 | Acorus calamus | MKI67 FHA domain-interacting nucleolar phosphoprotein-like | 633 | F:GO:0003723 | F:RNA binding | F:nucleic acid binding; F:RNA binding | view details | |||
| HO002052.1 | Acorus calamus | protein PMR5-like | 158 | C:GO:0016021 | C:integral component of membrane | F:transferase activity | view details | |||
| KY531680.1 | Acorus calamus | beta-galactosidase 8 isoform X1 | 2064 | P:GO:0005975; F:GO:0004565; C:GO:0005773; C:GO:0016021; C:GO:0048046 | P:carbohydrate metabolic process; F:beta-galactosidase activity; C:vacuole; C:integral component of membrane; C:apoplast | Beta-galactosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:beta-galactosidase activity | view details | ||
| KM399828.1 | Acorus calamus | GTF2H2 | 1164 | P:GO:0006289; P:GO:0006357; F:GO:0008270; C:GO:0000439; C:GO:0005675 | P:nucleotide-excision repair; P:regulation of transcription by RNA polymerase II; F:zinc ion binding; C:transcription factor TFIIH core complex; C:transcription factor TFIIH holo complex | P:DNA repair; P:nucleotide-excision repair; P:transcription, DNA-templated; F:zinc ion binding; C:transcription factor TFIIH core complex | view details | |||
| KY525911.1 | Acorus calamus | endonuclease MutS2 | 2016 | P:GO:0090304; F:GO:0000166; F:GO:0008094; F:GO:0016787; F:GO:0030983; C:GO:0032300 | P:nucleic acid metabolic process; F:nucleotide binding; F:ATP-dependent activity, acting on DNA; F:hydrolase activity; F:mismatched DNA binding; C:mismatch repair complex | Hydrolases | P:mismatch repair; P:negative regulation of DNA recombination; F:endonuclease activity; F:ATP binding; F:ATP hydrolysis activity; F:mismatched DNA binding | view details | ||
| AF193944.1 | Acorus calamus | cytochrome c oxidase subunit 1 | 1413 | P:GO:0006119; P:GO:0015990; P:GO:0022900; F:GO:0004129; F:GO:0020037; F:GO:0046872; C:GO:0005743; C:GO:0016021; C:GO:0045277 | P:oxidative phosphorylation; P:electron transport coupled proton transport; P:electron transport chain; F:cytochrome-c oxidase activity; F:heme binding; F:metal ion binding; C:mitochondrial inner membrane; C:integral component of membrane; C:respiratory chain complex IV | Acting on a heme group of donors; Cytochrome-c oxidase | P:aerobic respiration; F:cytochrome-c oxidase activity; F:heme binding; C:integral component of membrane; C:respiratory chain complex IV | view details | ||
| KM400103.1 | Acorus calamus | ankyrin repeat family protein | 924 | P:GO:0035246; F:GO:0016274; C:GO:0005634; C:GO:0005737 | P:peptidyl-arginine N-methylation; F:protein-arginine N-methyltransferase activity; C:nucleus; C:cytoplasm | Transferring one-carbon groups | F:protein binding; F:protein-arginine N-methyltransferase activity | view details | ||
| KY534881.1 | Acorus calamus | protein NRT1/ PTR FAMILY 6.1 | 1764 | P:GO:0006857; P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:oligopeptide transport; P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| AF039256.1 | Acorus calamus | ATPase subunit 1 | 1263 | P:GO:0015986; F:GO:0005524; F:GO:0043531; F:GO:0046933; C:GO:0005743; C:GO:0045261 | P:ATP synthesis coupled proton transport; F:ATP binding; F:ADP binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:mitochondrial inner membrane; C:proton-transporting ATP synthase complex, catalytic core F(1) | H(+)-transporting two-sector ATPase; Ligases | P:ATP synthesis coupled proton transport; P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding; F:adenyl ribonucleotide binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:proton-transporting ATP synthase complex, catalytic core F(1) | view details | ||
| KY541278.1 | Acorus calamus | denticleless protein homolog | 366 | P:GO:0007095; P:GO:0043161; F:GO:0030674; C:GO:0005634 | P:mitotic G2 DNA damage checkpoint signaling; P:proteasome-mediated ubiquitin-dependent protein catabolic process; F:protein-macromolecule adaptor activity; C:nucleus | F:protein binding | view details | |||
| KY526349.1 | Acorus calamus | PREDICTED: uncharacterized protein LOC104893382 | 423 | C:GO:0005637; C:GO:0016021 | C:nuclear inner membrane; C:integral component of membrane | no GO terms | view details | |||
| KM398314.1 | Acorus calamus | bushy growth protein | 681 | P:GO:0006338; P:GO:0006357; F:GO:0003712; C:GO:0000228; C:GO:0070603 | P:chromatin remodeling; P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:nuclear chromosome; C:SWI/SNF superfamily-type complex | P:chromatin remodeling; C:nuclear chromosome | view details | |||
| KM399229.1 | Acorus calamus | FMN-linked oxidoreductases superfamily protein | 1560 | P:GO:0002943; F:GO:0003677; F:GO:0017150; F:GO:0046872; F:GO:0050660 | P:tRNA dihydrouridine synthesis; F:DNA binding; F:tRNA dihydrouridine synthase activity; F:metal ion binding; F:flavin adenine dinucleotide binding | Acting on the CH-CH group of donors | F:catalytic activity; F:metal ion binding | view details | ||
| HM775144.1 | Acorus calamus | CYC-like protein 1 | 405 | P:GO:0006355; P:GO:2000032; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:regulation of secondary shoot formation; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| MN481074.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JN090105.1 | Acorus calamus | maturase K | 748 | P:GO:0006397; P:GO:0008033; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| LC219302.1 | Acorus calamus | maturase K | 789 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| MN481062.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 654 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| LC219079.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY526020.1 | Acorus calamus | vacuolar protein sorting-associated protein 25 | 534 | P:GO:0043328; F:GO:0003677; F:GO:0005198; F:GO:0008168; F:GO:0042803; C:GO:0000814 | P:protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway; F:DNA binding; F:structural molecule activity; F:methyltransferase activity; F:protein homodimerization activity; C:ESCRT II complex | Transferring one-carbon groups | P:multivesicular body sorting pathway; C:ESCRT II complex | view details | ||
| KM397878.1 | Acorus calamus | tetratricopeptide repeat-like superfamily protein | 1410 | F:protein binding | view details | |||||
| EU814656.1 | Acorus calamus | maturase K | 1028 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| AM890044.1 | Acorus calamus | photosystem II protein Z | 115 | P:GO:0042549; C:GO:0009535; C:GO:0009539; C:GO:0016021 | P:photosystem II stabilization; C:chloroplast thylakoid membrane; C:photosystem II reaction center; C:integral component of membrane | no IPS match | view details | |||
| JF940681.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 570 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KM823794.1 | Acorus calamus | RHOMBOID-like protein 3 isoform X1 | 454 | P:GO:0006508; F:GO:0004252; C:GO:0016021 | P:proteolysis; F:serine-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type endopeptidase activity; C:integral component of membrane | view details | ||
| HO002064.1 | Acorus calamus | cytochrome P450-like TBP protein | 159 | no GO terms | view details | |||||
| KM397550.1 | Acorus calamus | MLH1 | 2052 | P:GO:0006298; F:GO:0005524; F:GO:0016887; F:GO:0030983; C:GO:0005634; C:GO:0032300 | P:mismatch repair; F:ATP binding; F:ATP hydrolysis activity; F:mismatched DNA binding; C:nucleus; C:mismatch repair complex | Acting on acid anhydrides | P:mismatch repair; F:ATP binding; F:ATP hydrolysis activity; F:mismatched DNA binding; C:mismatch repair complex | view details | ||
| GQ435779.1 | Acorus calamus | cytochrome c heme attachment protein | 376 | P:GO:0017004; F:GO:0020037; C:GO:0009535; C:GO:0016021 | P:cytochrome complex assembly; F:heme binding; C:chloroplast thylakoid membrane; C:integral component of membrane | P:cytochrome complex assembly; F:heme binding | view details | |||
| HO002059.1 | Acorus calamus | Regulator of rDNA transcription protein 15 | 149 | no GO terms | view details | |||||
| FJ875010.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KY539767.1 | Acorus calamus | translin-associated protein X | 801 | P:GO:0090502; F:GO:0003723; F:GO:0004521; F:GO:0043565; C:GO:0005634; C:GO:0005737 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA binding; F:endoribonuclease activity; F:sequence-specific DNA binding; C:nucleus; C:cytoplasm | Acting on ester bonds | F:sequence-specific DNA binding | view details | ||
| HM775145.1 | Acorus calamus | CYC-like protein 1 | 399 | P:GO:0006355; F:GO:0003700 | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity | no IPS match | view details | |||
| FJ875014.1 | Acorus calamus | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1264 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| GQ248715.1 | Acorus calamus | DNA-directed RNA polymerase subunit beta | 385 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0032549; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding | view details | ||
| KY532256.1 | Acorus calamus | mRNA cap guanine-N7 methyltransferase 2 | 774 | P:GO:0006370; P:GO:0106005; F:GO:0003723; F:GO:0004482 | P:7-methylguanosine mRNA capping; P:RNA 5-cap (guanine-N7)-methylation; F:RNA binding; F:mRNA (guanine-N7-)-methyltransferase activity | mRNA (guanine-N(7)-)-methyltransferase | F:mRNA (guanine-N7-)-methyltransferase activity | view details | ||
| AM889946.1 | Acorus calamus | RNA polymerase beta' subunit | 485 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0008270; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:zinc ion binding; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| KY526397.1 | Acorus calamus | tRNase Z TRZ2, chloroplastic | 903 | P:GO:0034414; F:GO:0042781 | P:tRNA 3-trailer cleavage, endonucleolytic; F:3-tRNA processing endoribonuclease activity | Acting on ester bonds; Ribonuclease Z | no GO terms | view details | ||
| KY539659.1 | Acorus calamus | exonuclease mut-7 homolog isoform X2 | 1416 | P:GO:0006139; F:GO:0008408 | P:nucleobase-containing compound metabolic process; F:3-5 exonuclease activity | Acting on ester bonds | P:nucleobase-containing compound metabolic process; F:nucleic acid binding; F:3-5 exonuclease activity | view details | ||
| AM889770.1 | Acorus calamus | RNA polymerase beta subunit | 335 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0032549; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding | view details | ||
| KX365262.1 | Acorus calamus | RNA polymerase C | 454 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| LC387637.1 | Acorus calamus | S-adenosyl-L-methionine:3-hydroxy-N-methylcoclaurine 4-O-methyltransferase | 1068 | P:GO:0032259; F:GO:0008171; F:GO:0008757; F:GO:0046983 | P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| DQ407038.1 | Acorus calamus | RNA polymerase beta' subunit | 2516 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0008270; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:zinc ion binding; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| KM400211.1 | Acorus calamus | S-adenosyl-L-methionine-dependent methyltransferase superfamily protein | 765 | P:GO:0006370; P:GO:0106005; F:GO:0003723; F:GO:0004482 | P:7-methylguanosine mRNA capping; P:RNA 5-cap (guanine-N7)-methylation; F:RNA binding; F:mRNA (guanine-N7-)-methyltransferase activity | mRNA (guanine-N(7)-)-methyltransferase | F:mRNA (guanine-N7-)-methyltransferase activity | view details | ||
| FJ416892.1 | Acorus calamus | DNA-directed RNA polymerase III subunit 1 | 1632 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0046872; C:GO:0005666; C:GO:0005737 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:metal ion binding; C:RNA polymerase III complex; C:cytoplasm | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| GQ248876.1 | Acorus calamus | RNA polymerase C | 487 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| KM399395.1 | Acorus calamus | 15-cis-zeta-carotene isomerase, chloroplastic | 867 | P:GO:0016120; F:GO:0090471; C:GO:0016021 | P:carotene biosynthetic process; F:9,15,9-tri-cis-zeta-carotene isomerase activity; C:integral component of membrane | Cis-trans-isomerases | no GO terms | view details | ||
| AM889855.1 | Acorus calamus | RNA polymerase C | 549 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| KJ749886.1 | Acorus calamus | RNA polymerase beta subunit protein | 442 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| KY534172.1 | Acorus calamus | polyadenylate-binding protein RBP47B | 1260 | F:GO:0003729; C:GO:0005829 | F:mRNA binding; C:cytosol | F:nucleic acid binding; F:RNA binding | view details | |||
| GQ436214.1 | Acorus calamus | ---NA--- | 487 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | |||||
| KY537045.1 | Acorus calamus | protein GLE1 | 1854 | P:GO:0051028; F:GO:0005488; C:GO:0005634 | P:mRNA transport; F:binding; C:nucleus | P:poly(A)+ mRNA export from nucleus; C:nuclear pore | view details | |||
| AF123771.1 | Acorus calamus | NADH-plastoquinone oxidoreductase subunit 2 | 3668 | P:GO:0019684; P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:photosynthesis, light reaction; P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | no GO terms | view details | ||
| AF123843.1 | Acorus calamus | photosystem II CP47 chlorophyll apoprotein | 2193 | P:GO:0009772; P:GO:0018298; F:GO:0016168; F:GO:0045156; C:GO:0009523; C:GO:0009535; C:GO:0016021 | P:photosynthetic electron transport in photosystem II; P:protein-chromophore linkage; F:chlorophyll binding; F:electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity; C:photosystem II; C:chloroplast thylakoid membrane; C:integral component of membrane | Oxidoreductases | P:photosynthetic electron transport chain; P:photosynthetic electron transport in photosystem II; P:photosynthesis; P:photosynthesis, light reaction; F:chlorophyll binding; F:electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity; C:photosystem; C:photosystem II; C:membrane | view details | ||
| KY537293.1 | Acorus calamus | pre-rRNA-processing protein ESF1 | 1419 | P:GO:0006364; F:GO:0003723; C:GO:0005730 | P:rRNA processing; F:RNA binding; C:nucleolus | P:rRNA processing; C:nucleus | view details | |||
| KY525128.1 | Acorus calamus | Bacterial periplasmic spermidine/putrescine-binding protein | 1548 | P:GO:0009657; P:GO:0015846; F:GO:0019808; C:GO:0009507; C:GO:0042597 | P:plastid organization; P:polyamine transport; F:polyamine binding; C:chloroplast; C:periplasmic space | P:polyamine transport; F:polyamine binding; C:periplasmic space | view details | |||
| DQ005577.1 | Acorus calamus | PISTILLATA-like protein | 706 | P:GO:0045944; F:GO:0000977; F:GO:0003700; F:GO:0046983; C:GO:0005634 | P:positive regulation of transcription by RNA polymerase II; F:RNA polymerase II transcription regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus | view details | |||
| AF123813.1 | Acorus calamus | photosystem II 44 kDa protein | 2181 | P:GO:0009772; P:GO:0018298; F:GO:0016168; F:GO:0045156; F:GO:0046872; C:GO:0009523; C:GO:0009535; C:GO:0016021 | P:photosynthetic electron transport in photosystem II; P:protein-chromophore linkage; F:chlorophyll binding; F:electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity; F:metal ion binding; C:photosystem II; C:chloroplast thylakoid membrane; C:integral component of membrane | Oxidoreductases | P:photosynthetic electron transport chain; P:photosynthetic electron transport in photosystem II; P:photosynthesis; P:photosynthesis, light reaction; F:chlorophyll binding; F:electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity; C:photosystem; C:photosystem II; C:membrane | view details | ||
| NC_007407.1 | Acorus calamus | 153821 | F:ATP binding; F:ATPase | view details | ||||||
| KY537617.1 | Acorus calamus | hypothetical protein IFM89_026810 | 1980 | P:GO:0043087 | P:regulation of GTPase activity | no GO terms | view details | |||
| KY535187.1 | Acorus calamus | probable plastid-lipid-associated protein 12, chloroplastic | 1191 | no GO terms | view details | |||||
| AY812747.1 | Acorus calamus | hypothetical protein SADUNF_Sadunf17G0028900 | 713 | no GO terms | view details | |||||
| KY530568.1 | Acorus calamus | DUF3685 domain-containing protein | 1545 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| KM823796.1 | Acorus calamus | rhomboid-like protein 11, chloroplastic | 963 | P:GO:0006508; P:GO:0010142; P:GO:0016310; P:GO:0019287; P:GO:0080140; F:GO:0004252; F:GO:0004631; F:GO:0005524; F:GO:0019904; C:GO:0005777; C:GO:0009706; C:GO:0016021 | P:proteolysis; P:farnesyl diphosphate biosynthetic process, mevalonate pathway; P:phosphorylation; P:isopentenyl diphosphate biosynthetic process, mevalonate pathway; P:regulation of jasmonic acid metabolic process; F:serine-type endopeptidase activity; F:phosphomevalonate kinase activity; F:ATP binding; F:protein domain specific binding; C:peroxisome; C:chloroplast inner membrane; C:integral component of membrane | Acting on peptide bonds (peptidases); Phosphomevalonate kinase | F:serine-type endopeptidase activity; C:integral component of membrane | view details | ||
| KM398577.1 | Acorus calamus | ARC6 | 1626 | P:GO:0009739; P:GO:0010020; F:GO:0042803; F:GO:0043621; C:GO:0031357 | P:response to gibberellin; P:chloroplast fission; F:protein homodimerization activity; F:protein self-association; C:integral component of chloroplast inner membrane | no GO terms | view details | |||
| KM823799.1 | Acorus calamus | RHOMBOID-like protein 12, mitochondrial | 1113 | P:GO:0006508; F:GO:0004252; C:GO:0016021 | P:proteolysis; F:serine-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | F:serine-type endopeptidase activity; C:integral component of membrane | view details | ||
| KM397496.1 | Acorus calamus | DNA mismatch repair protein | 1851 | P:GO:0006298; P:GO:0090305; F:GO:0004519; F:GO:0005524; F:GO:0030983; C:GO:0009507 | P:mismatch repair; P:nucleic acid phosphodiester bond hydrolysis; F:endonuclease activity; F:ATP binding; F:mismatched DNA binding; C:chloroplast | Acting on ester bonds | P:mismatch repair; F:ATP binding; F:mismatched DNA binding | view details | ||
| AB017421.1 | Acorus calamus | ---NA--- | 713 | no IPS match | view details | |||||
| AB331311.1 | Acorus calamus | ribosomal protein L22 | 1239 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0009507; C:GO:0015934; C:GO:0015935 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:chloroplast; C:large ribosomal subunit; C:small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:large ribosomal subunit | view details | |||
| KY530063.1 | Acorus calamus | hypothetical protein CKAN_00302900 | 1296 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| HE651036.1 | Acorus calamus | maturase K | 2683 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | no IPS match | view details | |||
| AJ879453.1 | Acorus calamus | 153821 | F:ATP binding; F:ATPase | view details | ||||||
| AB017423.1 | Acorus calamus | ---NA--- | 713 | no IPS match | view details | |||||
| HE651032.1 | Acorus calamus | maturase K | 2683 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | no IPS match | view details | |||
| AB017422.1 | Acorus calamus | ---NA--- | 713 | no IPS match | view details | |||||
| AJ006146.1 | Acorus calamus | cytochrome c oxidase subunit II | 1400 | P:GO:0022900; P:GO:1902600; F:GO:0004129; F:GO:0005507; C:GO:0005743; C:GO:0016021; C:GO:0070469 | P:electron transport chain; P:proton transmembrane transport; F:cytochrome-c oxidase activity; F:copper ion binding; C:mitochondrial inner membrane; C:integral component of membrane; C:respirasome | Acting on a heme group of donors; Cytochrome-c oxidase | F:cytochrome-c oxidase activity; F:copper ion binding; C:membrane | view details | ||
| MT755635.1 | Acorus calamus | 153453 | F:ATP binding; F:ATPase | view details | ||||||
| AB017424.1 | Acorus calamus | ---NA--- | 713 | no IPS match | view details | |||||
| KM823795.1 | Acorus calamus | RHOMBOID-like protein 10, chloroplastic | 810 | P:GO:0006508; P:GO:0044255; F:GO:0004252; C:GO:0016021; C:GO:0019866; C:GO:0031969 | P:proteolysis; P:cellular lipid metabolic process; F:serine-type endopeptidase activity; C:integral component of membrane; C:organelle inner membrane; C:chloroplast membrane | Acting on peptide bonds (peptidases) | F:serine-type endopeptidase activity; C:integral component of membrane | view details |
lncRNA Information
Gene Information
| Gene ID | Species Name | Symbol | Aliases | Description | Start position | End position | Orientation | Genomic Context |
|---|---|---|---|---|---|---|---|---|
| 3677518 | Acorus calamus | ndhB | AccaCp068, ndh2 | NADH dehydrogenase subunit 2 | 93846 | 96078 | minus | |
| 3677517 | Acorus calamus | ndhB | AccaCp085, ndh2 | NADH dehydrogenase subunit 2 | 141893 | 144125 | plus | |
| 3677516 | Acorus calamus | ndhA | AccaCp078, ndh1 | NADH dehydrogenase subunit 1 | 119490 | 121692 | minus | |
| 3677515 | Acorus calamus | matK | AccaCp003 | maturase K | 2008 | 3543 | minus | |
| 3677514 | Acorus calamus | psbZ | AccaCp019 | photosystem II reaction center Z protein | 36442 | 36630 | plus | |
| 3677513 | Acorus calamus | infA | AccaCp058 | translation initiation factor 1 | 79502 | 79735 | minus | |
| 3677512 | Acorus calamus | clpP | AccaCp048 | ATP-dependent Clp protease proteolytic subunit | 69750 | 71762 | minus | |
| 3677511 | Acorus calamus | cemA | AccaCp033, ycf10 | heme-binding protein | 60998 | 61687 | plus | |
| 3677510 | Acorus calamus | ccsA | AccaCp072, ycf5 | cytochrome c biogenesis protein | 114299 | 115264 | plus | |
| 3677509 | Acorus calamus | atpI | AccaCp010 | ATPase IV subunit | 14798 | 15541 | minus | |
| 3677508 | Acorus calamus | atpH | AccaCp009 | ATPase III subunit | 13624 | 13869 | minus | |
| 3677507 | Acorus calamus | atpF | AccaCp008 | ATPase subunit I | 11855 | 13127 | minus | |
| 3677506 | Acorus calamus | atpE | AccaCp028 | ATP synthase epsilon chain | 54000 | 54404 | minus | |
| 3677505 | Acorus calamus | atpB | AccaCp029 | ATPase beta chain | 54401 | 55897 | minus | |
| 3677504 | Acorus calamus | atpA | AccaCp007 | ATP synthase CF1 alpha chain | 10264 | 11787 | minus | |
| 3677499 | Acorus calamus | psaJ | AccaCp041 | photosystem I subunit IX | 66920 | 67048 | plus | |
| 3677498 | Acorus calamus | psaI | AccaCp031 | photosystem I subunit VIII | 59416 | 59526 | plus | |
| 3677497 | Acorus calamus | psaC | AccaCp074 | photosystem I iron-sulfur center | 117109 | 117354 | minus | |
| 3677496 | Acorus calamus | psaB | AccaCp021 | PSI P700 apoprotein A2 | 37793 | 39997 | minus | |
| 3677495 | Acorus calamus | psaA | AccaCp022 | PSI P700 apoprotein A1 | 40023 | 42275 | minus | |
| 3677494 | Acorus calamus | petN | AccaCp015, ycf6 | cytochrome b6/f complex subunit N | 29037 | 29126 | plus | |
| 3677493 | Acorus calamus | petL | AccaCp039 | cytochrome b6/f complex subunit VI | 65765 | 65860 | plus | |
| 3677492 | Acorus calamus | petG | AccaCp040 | cytochrome B6-F complex subunit 5 | 66023 | 66136 | plus | |
| 3677491 | Acorus calamus | petD | AccaCp054 | cytochrome b6/f complex subunit 4 | 76280 | 77488 | plus | |
| 3677490 | Acorus calamus | petB | AccaCp053 | cytochrome b6 | 74658 | 76089 | plus | |
| 3677489 | Acorus calamus | petA | AccaCp034 | apocytochrome f precursor | 61877 | 62839 | plus | |
| 3677488 | Acorus calamus | ndhK | AccaCp026 | NADH dehydrogenase 27 kDa subunit | 49851 | 50570 | minus | |
| 3677487 | Acorus calamus | ndhJ | AccaCp025 | NADH dehydrogenase 30 kDa subunit | 49299 | 49775 | minus | |
| 3677486 | Acorus calamus | ndhI | AccaCp077, frxB | NADH dehydrogenase 18 kDa subunit | 118873 | 119412 | minus | |
| 3677485 | Acorus calamus | ndhH | AccaCp079 | NADH dehydrogenase 49 kDa subunit | 121694 | 122875 | minus | |
| 3677484 | Acorus calamus | ndhG | AccaCp076 | NADH dehydrogenase subunit 6 | 117997 | 118530 | minus | |
| 3677483 | Acorus calamus | ndhF | AccaCp070, ndh5 | NADH dehydrogenase subunit 5 | 110030 | 112249 | minus | |
| 3677482 | Acorus calamus | ndhE | AccaCp075, ndh4L | NADH dehydrogenase subunit 4L | 117599 | 117904 | minus | |
| 3677481 | Acorus calamus | ndhD | AccaCp073, ndh4 | NADH dehydrogenase subunit 4 | 115492 | 116994 | minus | |
| 3677480 | Acorus calamus | rpl2 | AccaCp065 | 50S ribosomal protein L2 | 84181 | 85671 | minus | |
| 3677479 | Acorus calamus | rpl2 | AccaCp088 | 50S ribosomal protein L2 | 152300 | 153790 | plus | |
| 3677478 | Acorus calamus | rpl16 | AccaCp061 | ribosomal protein L16 | 80948 | 82294 | minus | |
| 3677477 | Acorus calamus | rpl14 | AccaCp060 | 50S ribosomal protein L14 | 80438 | 80806 | minus | |
| 3677476 | Acorus calamus | rbcL | AccaCp030 | RuBisCO large subunit | 56693 | 58156 | plus | |
| 3677475 | Acorus calamus | psbT | AccaCp050 | photosystem II protein T | 73897 | 74013 | plus | |
| 3677474 | Acorus calamus | psbN | AccaCp051 | photosystem II reaction center N protein | 74073 | 74204 | minus | |
| 3677473 | Acorus calamus | psbM | AccaCp016 | photosystem II protein M | 30106 | 30210 | minus | |
| 3677472 | Acorus calamus | psbL | AccaCp036 | PSII reaction center subunit XII | 63976 | 64092 | minus | |
| 3677471 | Acorus calamus | psbK | AccaCp005 | PSII K protein | 7311 | 7496 | plus | |
| 3677470 | Acorus calamus | psbJ | AccaCp035 | photosystem II reaction center subunit X | 63718 | 63840 | minus | |
| 3677469 | Acorus calamus | psbI | AccaCp006 | photosystem II protein I | 7931 | 8041 | plus | |
| 3677468 | Acorus calamus | psbH | AccaCp052 | photosystem II reaction center protein H | 74317 | 74538 | plus | |
| 3677467 | Acorus calamus | psbF | AccaCp037 | photosystem II reaction center subunit VI | 64115 | 64234 | minus | |
| 3677466 | Acorus calamus | psbE | AccaCp038 | cytochrome b559 alpha subunit | 64244 | 64495 | minus | |
| 3677465 | Acorus calamus | psbD | AccaCp017 | photosystem II protein D2 | 33319 | 34380 | plus | |
| 3677464 | Acorus calamus | psbC | AccaCp018 | PSII 43 kDa protein | 34286 | 35749 | plus | |
| 3677463 | Acorus calamus | psbB | AccaCp049 | photosystem II P680 chlorophyll A apoprotein | 72204 | 73730 | plus | |
| 3677462 | Acorus calamus | psbA | AccaCp001 | photosystem II protein D1 | 464 | 1525 | minus | |
| 3677461 | Acorus calamus | rps4 | AccaCp024 | ribosomal protein S4 | 46089 | 46694 | minus | |
| 3677460 | Acorus calamus | rps3 | AccaCp062 | 30S ribosomal protein S3 | 82455 | 83111 | minus | |
| 3677459 | Acorus calamus | rps2 | AccaCp011 | 30S ribosomal protein S2 | 15775 | 16485 | minus | |
| 3677458 | Acorus calamus | rps19 | AccaCp064 | 30S ribosomal protein S19 | 83630 | 83908 | minus | |
| 3677457 | Acorus calamus | rps18 | AccaCp043 | 30S ribosomal protein S18 | 67753 | 68058 | plus | |
| 3677456 | Acorus calamus | rps16 | AccaCp004 | ribosomal protein S16 | 4982 | 6063 | minus | |
| 3677455 | Acorus calamus | rps15 | AccaCp080 | ribosomal protein S15 | 122981 | 123253 | minus | |
| 3677454 | Acorus calamus | rps14 | AccaCp020 | 30S ribosomal protein S14 | 37353 | 37655 | minus | |
| 3677453 | Acorus calamus | rps11 | AccaCp056 | 30S ribosomal protein S11 | 78738 | 79154 | minus | |
| 3677452 | Acorus calamus | rpoC2 | AccaCp012 | RNA polymerase beta' chain | 16668 | 20816 | minus | |
| 3677450 | Acorus calamus | rpoB | AccaCp014 | RNA polymerase beta chain | 23828 | 27079 | minus | |
| 3677449 | Acorus calamus | rpoA | AccaCp055 | RNA polymerase alpha chain | 77656 | 78669 | minus | |
| 3677448 | Acorus calamus | rpl36 | AccaCp057, secX | 50S ribosomal protein L36 | 79275 | 79388 | minus | |
| 3677447 | Acorus calamus | rpl33 | AccaCp042 | ribosomal protein L33 | 67413 | 67613 | plus | |
| 3677446 | Acorus calamus | rpl32 | AccaCp071 | ribosomal protein L32 | 113101 | 113274 | plus | |
| 3677445 | Acorus calamus | rpl23 | AccaCp066 | 50S ribosomal protein L23 | 85690 | 85971 | minus | |
| 3677444 | Acorus calamus | rpl23 | AccaCp087 | 50S ribosomal protein L23 | 152000 | 152281 | plus | |
| 3677443 | Acorus calamus | rpl22 | AccaCp063 | ribosomal protein L22 | 83197 | 83580 | minus | |
| 3677442 | Acorus calamus | rpl20 | AccaCp044 | 50S ribosomal protein L20 | 68326 | 68679 | minus | |
| 3677425 | Acorus calamus | rps8 | AccaCp059 | 30S ribosomal protein S8 | 79867 | 80265 | minus | |
| 3677424 | Acorus calamus | rps7 | AccaCp069 | 30S ribosomal protein S7 | 96396 | 96863 | minus | |
| 3677423 | Acorus calamus | rps7 | AccaCp084 | 30S ribosomal protein S7 | 141108 | 141575 | plus | |
| 3677401 | Acorus calamus | rps12 | AccaCp047 | ribosomal protein S12 | 69422 | 69535 | minus | |
| 3677401 | Acorus calamus | rps12 | AccaCp047 | ribosomal protein S12 | 96916 | 97709 | minus | |
| 3677400 | Acorus calamus | rps12 | AccaCp046 | ribosomal protein S12 | 69422 | 69535 | minus | |
| 3677400 | Acorus calamus | rps12 | AccaCp046 | ribosomal protein S12 | 140262 | 141055 | plus | |
| 3677399 | Acorus calamus | ycf4 | AccaCp032 | photosystem I assembly protein Ycf4 | 59902 | 60456 | plus | |
| 3677398 | Acorus calamus | ycf3 | AccaCp023 | photosystem I assembly protein Ycf3 | 42894 | 44873 | minus | |
| 3677395 | Acorus calamus | ycf1 | AccaCp081 | Ycf1 | 123624 | 129365 | minus | |
| 3677389 | Acorus calamus | ndhC | AccaCp027, ndh3 | NADH dehydrogenase subunit 3 | 50624 | 50986 | minus | |
| 3677397 | Acorus calamus | ycf2 | AccaCp086 | Ycf2 | 145318 | 151548 | minus | |
| 3677396 | Acorus calamus | ycf2 | AccaCp067 | Ycf2 | 86423 | 92653 | plus | |
| 3677522 | Acorus calamus | 23S rRNA | AccaCr007 | 23S ribosomal RNA | 131412 | 134222 | minus | |
| 3677521 | Acorus calamus | 23S rRNA | AccaCr002 | 23S ribosomal RNA | 103749 | 106559 | plus | |
| 3677520 | Acorus calamus | 16S rRNA | AccaCr008 | 16S ribosomal RNA | 136642 | 138132 | minus | |
| 3677519 | Acorus calamus | 16S rRNA | AccaCr001 | 16S ribosomal RNA | 99839 | 101329 | plus | |
| 3677503 | Acorus calamus | 5S rRNA | AccaCr005 | 5S ribosomal RNA | 130873 | 130993 | minus | |
| 3677502 | Acorus calamus | 5S rRNA | AccaCr004 | 5S ribosomal RNA | 106978 | 107098 | plus | |
| 3677501 | Acorus calamus | 4.5S rRNA | AccaCr006 | 4.5S ribosomal RNA | 131211 | 131313 | minus | |
| 3677500 | Acorus calamus | 4.5S rRNA | AccaCr003 | 4.5S ribosomal RNA | 106658 | 106760 | plus | |
| 3677441 | Acorus calamus | tRNA-His (GUG) | AccaCt023 | tRNA | 84075 | 84152 | plus | |
| 3677440 | Acorus calamus | tRNA-Gly (UCC) | AccaCt005 | tRNA | 9171 | 10002 | plus | |
| 3677439 | Acorus calamus | tRNA-Gly (GCC) | AccaCt013 | tRNA | 36907 | 36977 | plus | |
| 3677438 | Acorus calamus | tRNA-Glu (UUC) | AccaCt010 | tRNA | 31292 | 31364 | minus | |
| 3677437 | Acorus calamus | tRNA-Gln (UUG) | AccaCt003 | tRNA | 6888 | 6960 | minus | |
| 3677436 | Acorus calamus | tRNA-fMet (CAU) | AccaCt014 | tRNA | 37118 | 37191 | minus | |
| 3677435 | Acorus calamus | tRNA-Cys (GCA) | AccaCt007 | tRNA | 28086 | 28156 | plus | |
| 3677434 | Acorus calamus | tRNA-Asp (GUC) | AccaCt008 | tRNA | 30734 | 30807 | minus | |
| 3677433 | Acorus calamus | tRNA-Asn (GUU) | AccaCt030 | tRNA | 107931 | 108002 | minus | |
| 3677432 | Acorus calamus | tRNA-Asn (GUU) | AccaCt032 | tRNA | 129969 | 130040 | plus | |
| 3677431 | Acorus calamus | tRNA-Arg (UCU) | AccaCt006 | tRNA | 10097 | 10168 | plus | |
| 3677430 | Acorus calamus | tRNA-Arg (ACG) | AccaCt033 | tRNA | 130538 | 130611 | minus | |
| 3677429 | Acorus calamus | tRNA-Arg (ACG) | AccaCt029 | tRNA | 107360 | 107433 | plus | |
| 3677428 | Acorus calamus | tRNA-Ala (UGC) | AccaCt034 | tRNA | 134375 | 135246 | minus | |
| 3677426 | Acorus calamus | tRNA-Ala (UGC) | AccaCt028 | tRNA | 102725 | 103596 | plus | |
| 3677422 | Acorus calamus | tRNA-Thr (UGU) | AccaCt016 | tRNA | 47045 | 47117 | minus | |
| 3677421 | Acorus calamus | tRNA-Thr (GGU) | AccaCt011 | tRNA | 31882 | 31953 | plus | |
| 3677420 | Acorus calamus | tRNA-Ser (UGA) | AccaCt012 | tRNA | 35969 | 36061 | minus | |
| 3677419 | Acorus calamus | tRNA-Ser (GGA) | AccaCt015 | tRNA | 45702 | 45789 | plus | |
| 3677418 | Acorus calamus | tRNA-Ser (GCU) | AccaCt004 | tRNA | 8158 | 8245 | minus | |
| 3677417 | Acorus calamus | tRNA-Pro (UGG) | AccaCt022 | tRNA | 66489 | 66562 | minus | |
| 3677416 | Acorus calamus | tRNA-Phe (GAA) | AccaCt018 | tRNA | 48841 | 48913 | plus | |
| 3677415 | Acorus calamus | tRNA-Met (CAU) | AccaCt020 | tRNA | 53763 | 53834 | plus | |
| 3677414 | Acorus calamus | tRNA-Lys (UUU) | AccaCt002 | tRNA | 1716 | 4287 | minus | |
| 3677412 | Acorus calamus | tRNA-Leu (UAG) | AccaCt031 | tRNA | 114138 | 114217 | plus | |
| 3677411 | Acorus calamus | tRNA-Leu (UAA) | AccaCt017 | tRNA | 47860 | 48470 | plus | |
| 3677410 | Acorus calamus | tRNA-Leu (CAA) | AccaCt025 | tRNA | 93202 | 93282 | minus | |
| 3677409 | Acorus calamus | tRNA-Leu (CAA) | AccaCt037 | tRNA | 144689 | 144769 | plus | |
| 3677408 | Acorus calamus | tRNA-Ile (GAU) | AccaCt035 | tRNA | 135315 | 136328 | minus | |
| 3677406 | Acorus calamus | tRNA-Ile (GAU) | AccaCt027 | tRNA | 101643 | 102656 | plus | |
| 3677405 | Acorus calamus | tRNA-Ile (CAU) | AccaCt024 | tRNA | 86138 | 86211 | minus | |
| 3677404 | Acorus calamus | tRNA-Ile (CAU) | AccaCt038 | tRNA | 151760 | 151833 | plus | |
| 3677403 | Acorus calamus | tRNA-His (GUG) | AccaCt001 | tRNA | ||||
| 3677394 | Acorus calamus | tRNA-Val (UAC) | AccaCt019 | tRNA | 52899 | 53579 | minus | |
| 3677393 | Acorus calamus | tRNA-Val (GAC) | AccaCt036 | tRNA | 138360 | 138431 | minus | |
| 3677392 | Acorus calamus | tRNA-Val (GAC) | AccaCt026 | tRNA | 99540 | 99611 | plus | |
| 3677391 | Acorus calamus | tRNA-Tyr (GUA) | AccaCt009 | tRNA | 31144 | 31227 | minus | |
| 3877511 | Acorus calamus | cemA | AccaCp033, ycf10 | heme-binding protein | 60998 | 61687 | plus | |
| 3877429 | Acorus calamus | tRNA-Arg (ACG) | AccaCt029 | tRNA | 107360 | 107433 | plus |
miRNA Information
| Acc. No. | Species Name | Sequence length | Total pre-miRNA | Sequence |
|---|---|---|---|---|
| DQ008852.1 | Acorus calamus | 982 | 13 Details |
|
| KM103412.1 | Acorus calamus | 1215 | 17 Details |
|
| DQ008654.1 | Acorus calamus | 2922 | 37 Details |
|
| AF203679.1 | Acorus calamus | 988 | 14 Details |
|
| KM397825.1 | Acorus calamus | 1012 | 8 Details |
RNA-Seq Information
| Experiment Acc. | Species Name | SRA Study | SRA Run | Treatment | Development Stage | Tissue | Data Accesss |
|---|---|---|---|---|---|---|---|
| SRX8123254 | Acorus calamus | SRP256576 | SRR11553473 | No | No | All diverse tissue | |
| SRX8123253 | Acorus calamus | SRS6487689 | SRR11553472 | No | No | All diverse tissue | |
| SRX8123252 | Acorus calamus | SRP256576 | SRR11553471 | No | No | All | |
| SRX8123233 | Acorus calamus | SRP256576 | SRR11553605 | No | No | All | |
| SRX5112834 | Acorus calamus | SRS4121727 | SRR8298303 | No | No | leaf | |
| SRX3469544 | Acorus calamus | SRS2756999 | SRR6374688 | No | Young leaves | leaves |
Other non-coding Sequence Information
* If data is not displayed please access the respective tab