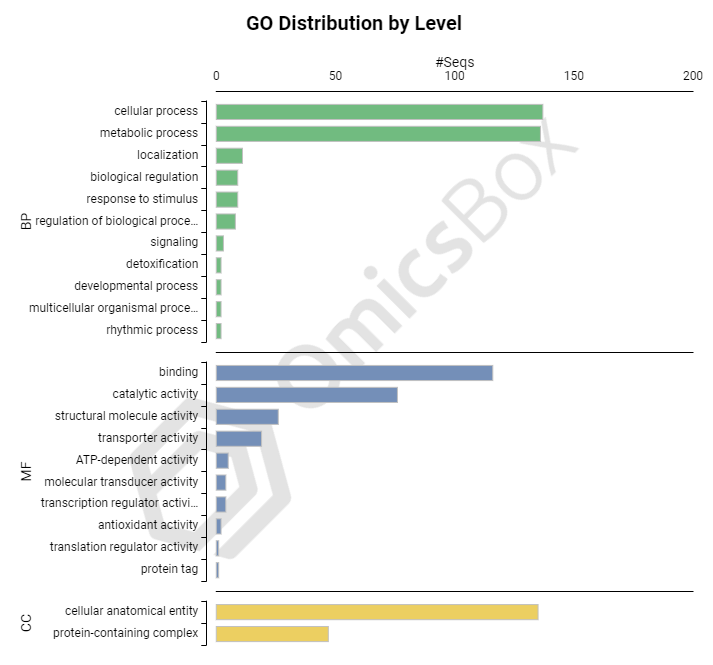
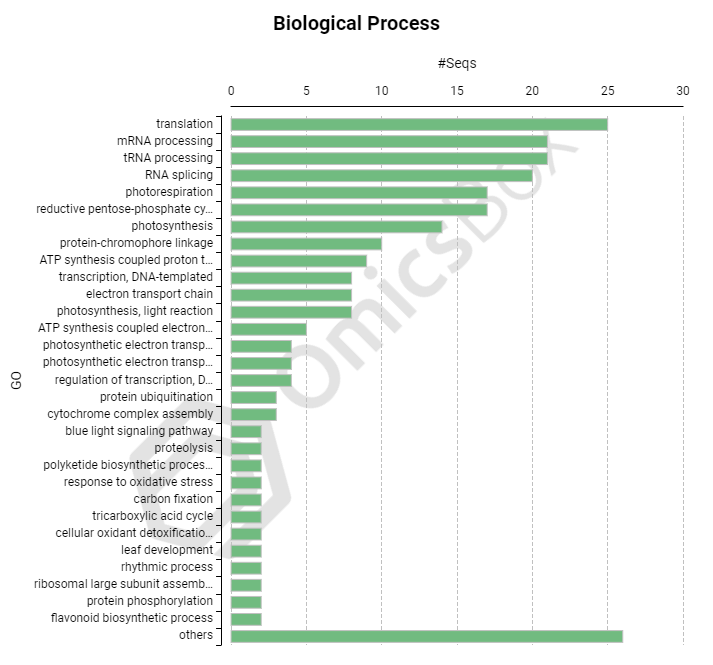
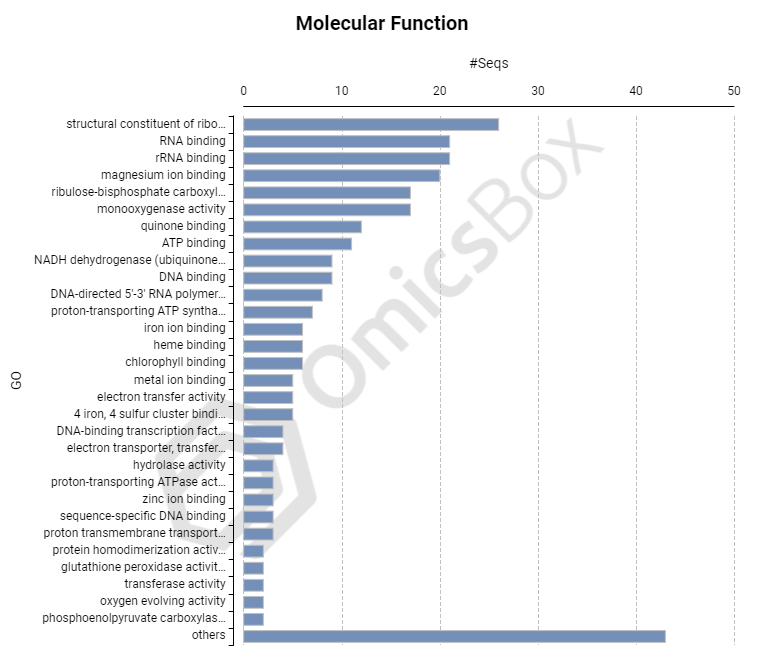
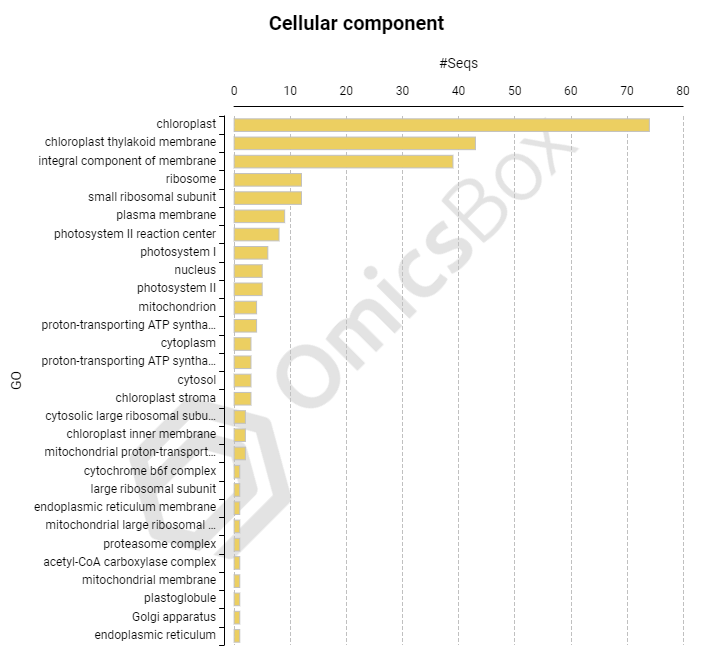
Coding Information
| Acc No. | Species Name | Description | Length | GO IDs | GO Names | Enzyme Names | InterPro GO Names | Peptide Information | BLAST | Nucleotide Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| GQ434218.1 | Datura innoxia | maturase K | 731 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| BQ047792.1 | Datura innoxia | phospholipase D beta 1-like | 559 | P:GO:0006355; P:GO:0009395; P:GO:0046470; F:GO:0003677; F:GO:0004630; F:GO:0005509; F:GO:0070290; C:GO:0005634; C:GO:0005886 | P:regulation of transcription, DNA-templated; P:phospholipid catabolic process; P:phosphatidylcholine metabolic process; F:DNA binding; F:phospholipase D activity; F:calcium ion binding; F:N-acylphosphatidylethanolamine-specific phospholipase D activity; C:nucleus; C:plasma membrane | Phospholipase D; Acting on ester bonds | F:catalytic activity | view details | ||
| FJ599222.1 | Datura innoxia | COSII_At3g55800 | 307 | P:GO:0005986; P:GO:0006000; P:GO:0006002; P:GO:0006094; P:GO:0030388; F:GO:0042132; C:GO:0005737 | P:sucrose biosynthetic process; P:fructose metabolic process; P:fructose 6-phosphate metabolic process; P:gluconeogenesis; P:fructose 1,6-bisphosphate metabolic process; F:fructose 1,6-bisphosphate 1-phosphatase activity; C:cytoplasm | Sugar-phosphatase; Fructose-bisphosphatase | no IPS match | view details | ||
| GQ274313.1 | Datura innoxia | maturase K | 771 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| X87826.1 | Datura innoxia | cellulose synthase-like protein D3 | 949 | P:GO:0009833; P:GO:0030244; P:GO:0097502; F:GO:0016760; F:GO:0051753; C:GO:0005886; C:GO:0016021 | P:plant-type primary cell wall biogenesis; P:cellulose biosynthetic process; P:mannosylation; F:cellulose synthase (UDP-forming) activity; F:mannan synthase activity; C:plasma membrane; C:integral component of membrane | Cellulose synthase (UDP-forming) | P:cellulose biosynthetic process; F:cellulose synthase (UDP-forming) activity; C:membrane | view details | ||
| MG218388.1 | Datura innoxia | Protein TAR1 | 351 | no IPS match | view details | |||||
| KJ676865.1 | Datura innoxia | tropinone reductase I | 822 | P:GO:0009710; F:GO:0050356; C:GO:0016021 | P:tropane alkaloid biosynthetic process; F:tropine dehydrogenase activity; C:integral component of membrane | Tropinone reductase I | F:oxidoreductase activity | view details | ||
| MH931115.1 | Datura innoxia | agamous-like MADS-box protein AGL8 homolog | 618 | P:GO:0045944; F:GO:0000978; F:GO:0000981; F:GO:0046983; C:GO:0005634 | P:positive regulation of transcription by RNA polymerase II; F:RNA polymerase II cis-regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity, RNA polymerase II-specific; F:protein dimerization activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus | view details | |||
| MH311559.1 | Datura innoxia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 603 | P:GO:0009853; P:GO:0015977; P:GO:0015979; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016491; F:GO:0016829; F:GO:0016984; F:GO:0046872; C:GO:0009507; C:GO:0009536 | P:photorespiration; P:carbon fixation; P:photosynthesis; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:oxidoreductase activity; F:lyase activity; F:ribulose-bisphosphate carboxylase activity; F:metal ion binding; C:chloroplast; C:plastid | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | |||
| MH931116.1 | Datura innoxia | FRUITFUL-like protein | 438 | P:GO:0006355; P:GO:0006357; P:GO:0045944; F:GO:0000977; F:GO:0000978; F:GO:0000981; F:GO:0003677; F:GO:0003700; F:GO:0046983; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:regulation of transcription by RNA polymerase II; P:positive regulation of transcription by RNA polymerase II; F:RNA polymerase II transcription regulatory region sequence-specific DNA binding; F:RNA polymerase II cis-regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity, RNA polymerase II-specific; F:DNA binding; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus | view details | |||
| JX996065.1 | Datura innoxia | maturase K | 870 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MG218484.1 | Datura innoxia | Protein TAR1 | 351 | no IPS match | view details | |||||
| FJ829038.1 | Datura innoxia | Integrase catalytic core | 278 | P:GO:0006260; P:GO:0006352; P:GO:0010020; P:GO:0015074; P:GO:0048285; P:GO:0050832; P:GO:0051301; F:GO:0000166; F:GO:0003676; F:GO:0003677; F:GO:0003689; F:GO:0003924; F:GO:0005524; F:GO:0005525; F:GO:0008270; F:GO:0009916; F:GO:0016491; F:GO:0046872; F:GO:0102721; C:GO:0005663; C:GO:0005737; C:GO:0009507; C:GO:0016020; C:GO:0016021; C:GO:0032153; C:GO:0070469 | P:DNA replication; P:DNA-templated transcription, initiation; P:chloroplast fission; P:DNA integration; P:organelle fission; P:defense response to fungus; P:cell division; F:nucleotide binding; F:nucleic acid binding; F:DNA binding; F:DNA clamp loader activity; F:GTPase activity; F:ATP binding; F:GTP binding; F:zinc ion binding; F:alternative oxidase activity; F:oxidoreductase activity; F:metal ion binding; F:ubiquinol:oxygen oxidoreductase activity; C:DNA replication factor C complex; C:cytoplasm; C:chloroplast; C:membrane; C:integral component of membrane; C:cell division site; C:respirasome | no GO terms | view details | |||
| MH285781.1 | Datura innoxia | maturase K | 772 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004713; F:GO:0004715; F:GO:0016740; C:GO:0005737; C:GO:0009507; C:GO:0009536; C:GO:0043231 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:protein tyrosine kinase activity; F:non-membrane spanning protein tyrosine kinase activity; F:transferase activity; C:cytoplasm; C:chloroplast; C:plastid; C:intracellular membrane-bounded organelle | P:mRNA processing; C:chloroplast | view details | |||
| MK519991.1 | Datura innoxia | maturase K | 785 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MH931117.1 | Datura innoxia | FRUITFUL-like protein | 438 | P:GO:0045944; F:GO:0000978; F:GO:0000981; F:GO:0046983; C:GO:0005634 | P:positive regulation of transcription by RNA polymerase II; F:RNA polymerase II cis-regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity, RNA polymerase II-specific; F:protein dimerization activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus | view details | |||
| MW374693.1 | Datura innoxia | 360 | no IPS match | view details | ||||||
| MH717196.1 | Datura innoxia | maturase K | 784 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004713; F:GO:0004715; F:GO:0016740; C:GO:0005737; C:GO:0009507; C:GO:0009536; C:GO:0043231 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:protein tyrosine kinase activity; F:non-membrane spanning protein tyrosine kinase activity; F:transferase activity; C:cytoplasm; C:chloroplast; C:plastid; C:intracellular membrane-bounded organelle | P:mRNA processing; C:chloroplast | view details | |||
| JN796937.1 | Datura innoxia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 666 | P:GO:0009853; P:GO:0015977; P:GO:0015979; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016491; F:GO:0016829; F:GO:0016984; F:GO:0046872; C:GO:0009507; C:GO:0009536 | P:photorespiration; P:carbon fixation; P:photosynthesis; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:oxidoreductase activity; F:lyase activity; F:ribulose-bisphosphate carboxylase activity; F:metal ion binding; C:chloroplast; C:plastid | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | |||
| KC146596.1 | Datura innoxia | maturase K | 789 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004713; F:GO:0004715; F:GO:0016740; C:GO:0005737; C:GO:0009507; C:GO:0009536; C:GO:0043231 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:protein tyrosine kinase activity; F:non-membrane spanning protein tyrosine kinase activity; F:transferase activity; C:cytoplasm; C:chloroplast; C:plastid; C:intracellular membrane-bounded organelle | P:mRNA processing; C:chloroplast | view details | |||
| FJ829041.1 | Datura innoxia | reverse transcriptase | 292 | P:GO:0006278; P:GO:0090502; F:GO:0003676; F:GO:0003964; F:GO:0004523; F:GO:0016740; F:GO:0016779 | P:RNA-dependent DNA biosynthetic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:nucleic acid binding; F:RNA-directed DNA polymerase activity; F:RNA-DNA hybrid ribonuclease activity; F:transferase activity; F:nucleotidyltransferase activity | no GO terms | view details | |||
| MH107772.1 | Datura innoxia | maturase K | 804 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004713; F:GO:0004715; F:GO:0016740; C:GO:0005737; C:GO:0009507; C:GO:0009536; C:GO:0043231 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:protein tyrosine kinase activity; F:non-membrane spanning protein tyrosine kinase activity; F:transferase activity; C:cytoplasm; C:chloroplast; C:plastid; C:intracellular membrane-bounded organelle | P:mRNA processing; C:chloroplast | view details | |||
| KF365991.1 | Datura innoxia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 582 | P:GO:0009853; P:GO:0015977; P:GO:0015979; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016491; F:GO:0016829; F:GO:0016984; F:GO:0046872; C:GO:0009507; C:GO:0009536 | P:photorespiration; P:carbon fixation; P:photosynthesis; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:oxidoreductase activity; F:lyase activity; F:ribulose-bisphosphate carboxylase activity; F:metal ion binding; C:chloroplast; C:plastid | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | |||
| AM177609.1 | Datura innoxia | putrescine N-methyltransferase | 1023 | P:GO:0006596; P:GO:0009446; P:GO:0032259; P:GO:0042179; F:GO:0003824; F:GO:0004766; F:GO:0008168; F:GO:0016740; F:GO:0030750 | P:polyamine biosynthetic process; P:putrescine biosynthetic process; P:methylation; P:nicotine biosynthetic process; F:catalytic activity; F:spermidine synthase activity; F:methyltransferase activity; F:transferase activity; F:putrescine N-methyltransferase activity | F:catalytic activity; F:putrescine N-methyltransferase activity | view details | |||
| JN244340.1 | Datura innoxia | maturase K | 731 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004713; F:GO:0004715; F:GO:0016740; C:GO:0005737; C:GO:0009507; C:GO:0009536; C:GO:0043231 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:protein tyrosine kinase activity; F:non-membrane spanning protein tyrosine kinase activity; F:transferase activity; C:cytoplasm; C:chloroplast; C:plastid; C:intracellular membrane-bounded organelle | P:mRNA processing; C:chloroplast | view details | |||
| AM233350.1 | Datura innoxia | ATP synthase beta subunit | 1008 | P:GO:0006754; P:GO:0006811; P:GO:0015986; P:GO:0042776; P:GO:0046034; P:GO:1902600; F:GO:0000166; F:GO:0005524; F:GO:0046933; F:GO:0046961; C:GO:0005739; C:GO:0005753; C:GO:0009507; C:GO:0009535; C:GO:0009536; C:GO:0009579; C:GO:0016020; C:GO:0031090; C:GO:0045261 | P:ATP biosynthetic process; P:ion transport; P:ATP synthesis coupled proton transport; P:mitochondrial ATP synthesis coupled proton transport; P:ATP metabolic process; P:proton transmembrane transport; F:nucleotide binding; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:mitochondrion; C:mitochondrial proton-transporting ATP synthase complex; C:chloroplast; C:chloroplast thylakoid membrane; C:plastid; C:thylakoid; C:membrane; C:organelle membrane; C:proton-transporting ATP synthase complex, catalytic core F(1) | P:ATP synthesis coupled proton transport; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:proton-transporting ATP synthase complex, catalytic core F(1) | view details | |||
| OK447742.1 | Datura innoxia | Protein TAR1 | 474 | no GO terms | view details | |||||
| MG222383.1 | Datura innoxia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0015977; P:GO:0015979; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016491; F:GO:0016829; F:GO:0016984; F:GO:0046872; C:GO:0009507; C:GO:0009536 | P:photorespiration; P:carbon fixation; P:photosynthesis; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:oxidoreductase activity; F:lyase activity; F:ribulose-bisphosphate carboxylase activity; F:metal ion binding; C:chloroplast; C:plastid | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | |||
| FJ829039.1 | Datura innoxia | 297 | no GO terms | view details | ||||||
| AY995183.1 | Datura innoxia | 1161 | P:lipid biosynthetic process; P:biosynthetic process; F:farnesyl-diphosphate farnesyltransferase activity; F:farnesyltranstransferase activity | view details | ||||||
| FJ829040.1 | Datura innoxia | 297 | no IPS match | view details | ||||||
| KU556644.1 | Datura innoxia | 621 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| MK525669.1 | Datura innoxia | 549 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| MG224012.1 | Datura innoxia | 552 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| JN244360.1 | Datura innoxia | 663 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| JX996059.1 | Datura innoxia | 1264 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| FJ829035.1 | Datura innoxia | 295 | no GO terms | view details | ||||||
| JN244359.1 | Datura innoxia | 663 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| KY050729.1 | Datura innoxia | 303 | P:translation; F:structural constituent of ribosome; C:ribosome | view details | ||||||
| FJ599142.1 | Datura innoxia | 311 | no IPS match | view details | ||||||
| JN244358.1 | Datura innoxia | 663 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| EF395184.1 | Datura innoxia | 579 | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; C:nucleus | view details | ||||||
| JN244339.1 | Datura innoxia | 731 | P:mRNA processing; C:chloroplast | view details | ||||||
| AM177610.1 | Datura innoxia | 1026 | F:catalytic activity; F:putrescine N-methyltransferase activity | view details | ||||||
| FJ829037.1 | Datura innoxia | 294 | no GO terms | view details | ||||||
| GQ274316.1 | Datura innoxia | 553 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| FJ829036.1 | Datura innoxia | 292 | no GO terms | view details | ||||||
| MH311571.1 | Datura innoxia | 603 | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||||||
| EF438895.1 | Datura innoxia | 932 | P:mRNA processing; C:chloroplast | view details | ||||||
| JN244341.1 | Datura innoxia | 731 | P:mRNA processing; C:chloroplast | view details | ||||||
| BQ047786.1 | Datura innoxia | 585 | no GO terms | view details | ||||||
| OK447743.1 | Datura innoxia | 471 | no GO terms | view details | ||||||
| AB188673.1 | Datura innoxia | 198 | no IPS match | view details | ||||||
| BQ047784.1 | Datura innoxia | 311 | no GO terms | view details | ||||||
| GQ436148.1 | Datura innoxia | RNA polymerase beta subunit | 487 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| GQ273984.1 | Datura innoxia | RpoB | 385 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0032549; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5 -3 RNA polymerase activity; F:ribonucleoside binding; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5 -3 RNA polymerase activity; F:ribonucleoside binding | view details | ||
| GQ273985.1 | Datura innoxia | RNA polymerase beta subunit | 487 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5 -3 RNA polymerase activity; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5 -3 RNA polymerase activity | view details | ||
| AY995182.1 | Datura innoxia | squalene monooxygenase-like | 1990 | P:GO:0009414; P:GO:0009723; P:GO:0016126; F:GO:0004506; F:GO:0050660; C:GO:0005783; C:GO:0016021 | P:response to water deprivation; P:response to ethylene; P:sterol biosynthetic process; F:squalene monooxygenase activity; F:flavin adenine dinucleotide binding; C:endoplasmic reticulum; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Squalene monooxygenase | P:sterol biosynthetic process; F:squalene monooxygenase activity; F:flavin adenine dinucleotide binding; C:integral component of membrane | view details | ||
| X87827.1 | Datura innoxia | dual specificity tyrosine-phosphorylation-regulated kinase 1B-like | 659 | P:GO:0009789; P:GO:0018105; P:GO:0018107; P:GO:0018108; P:GO:0046777; F:GO:0004674; F:GO:0004713; F:GO:0005524; C:GO:0005737 | P:positive regulation of abscisic acid-activated signaling pathway; P:peptidyl-serine phosphorylation; P:peptidyl-threonine phosphorylation; P:peptidyl-tyrosine phosphorylation; P:protein autophosphorylation; F:protein serine/threonine kinase activity; F:protein tyrosine kinase activity; F:ATP binding; C:cytoplasm | Transferring phosphorus-containing groups | no GO terms | view details | ||
| AY995181.1 | Datura innoxia | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 | 932 | P:GO:0008299; P:GO:0015936; P:GO:0016126; F:GO:0004420; C:GO:0005778; C:GO:0005789; C:GO:0016021 | P:isoprenoid biosynthetic process; P:coenzyme A metabolic process; P:sterol biosynthetic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; C:peroxisomal membrane; C:endoplasmic reticulum membrane; C:integral component of membrane | Hydroxymethylglutaryl-CoA reductase (NADPH) | P:isoprenoid biosynthetic process; P:coenzyme A metabolic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; F:protein binding; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| KC146611.1 | Datura innoxia | NADH dehydrogenase subunit | 2001 | P:GO:0042773; F:GO:0008137; F:GO:0048038; C:GO:0009535; C:GO:0016021 | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; C:chloroplast thylakoid membrane; C:integral component of membrane | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | P:ATP synthesis coupled electron transport; F:NADH dehydrogenase (ubiquinone) activity | view details |
lncRNA Information
Gene Information
| Gene ID | Species Name | Symbol | Aliases | Description | Start position | End position | Orientation | Genomic Context |
|---|---|---|---|---|---|---|---|---|
| 40527098 | Datura innoxia | AV2 | FLA66_gp1 | AV2 protein | 147 | 497 | plus | |
| 40527097 | Datura innoxia | AC2 | FLA66_gp4 | transcriptional activator protein | 1225 | 1632 | minus | |
| 40527096 | Datura innoxia | AC1 | FLA66_gp5 | replication-associated protein | 1562 | 2674 | minus | |
| 40527095 | Datura innoxia | AC4 | FLA66_gp6 | AC4 protein | 2206 | 2463 | minus | |
| 40527094 | Datura innoxia | AV1 | FLA66_gp2 | coat protein | 307 | 1083 | plus | |
| 40527093 | Datura innoxia | AC3 | FLA66_gp3 | replication enhancer protein | 1080 | 1484 | minus |
miRNA Information
| Acc. No. | Species Name | Sequence length | Total pre-miRNA | Sequence |
|---|---|---|---|---|
| BQ047785.1 | Datura innoxia | 198 | 2 Details |
RNA-Seq Information
| Experiment Acc. | Species Name | SRA Study | SRA Run | Treatment | Development Stage | Tissue | Data Accesss |
|---|---|---|---|---|---|---|---|
| SRX10578028 | Datura innoxia | SRP314413 | SRR14211379 | leaf | |||
| SRX10578027 | Datura innoxia | SRP314413 | SRR14211380 | root | |||
| SRX10578026 | Datura innoxia | SRP314413 | SRR14211381 | root |
Other non-coding Sequence Information
| Acc. No | Species Name | Length | GC content | Status | Sequence |
|---|---|---|---|---|---|
| FJ599241.1 | Datura innoxia | 606 | 39.54% | Non-coding | |
| EF590811.1 | Datura innoxia | 187 | 31.38% | Non-coding | |
| X87828.1 | Datura innoxia | 83 | 41.67% | Non-coding |
* If data is not displayed please access the respective tab