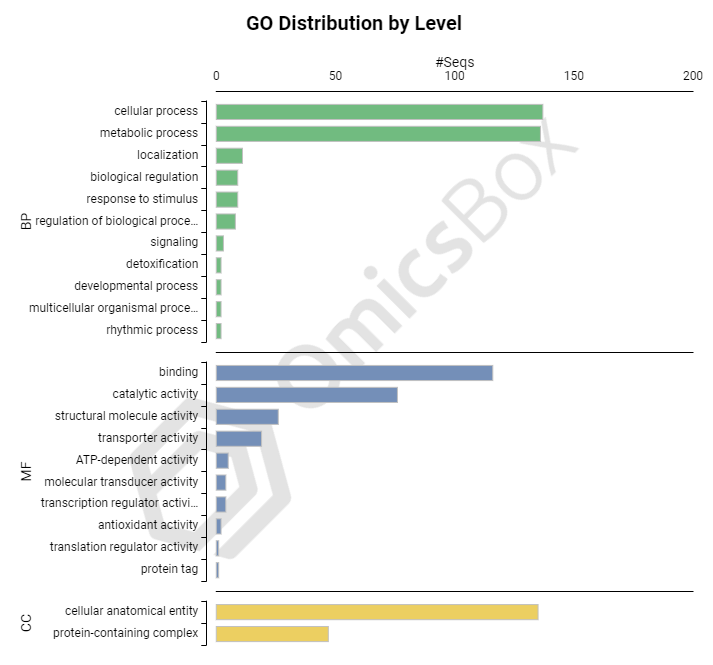
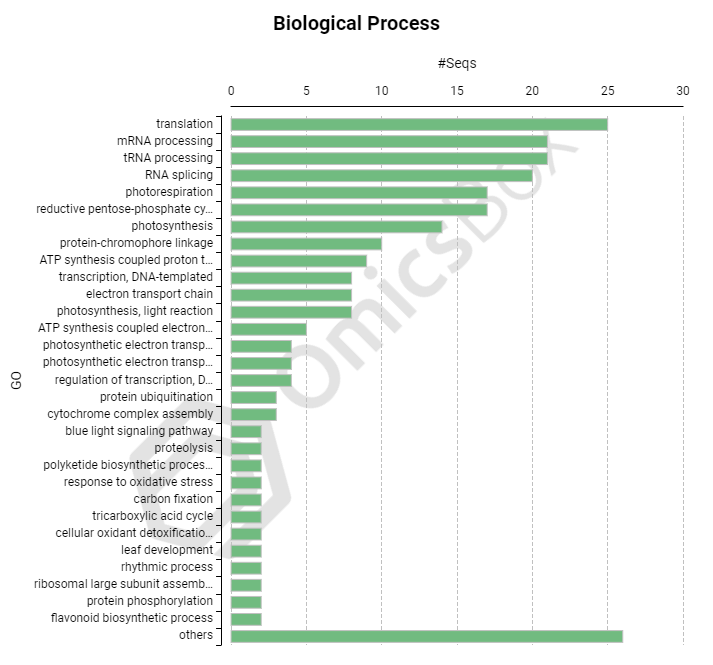
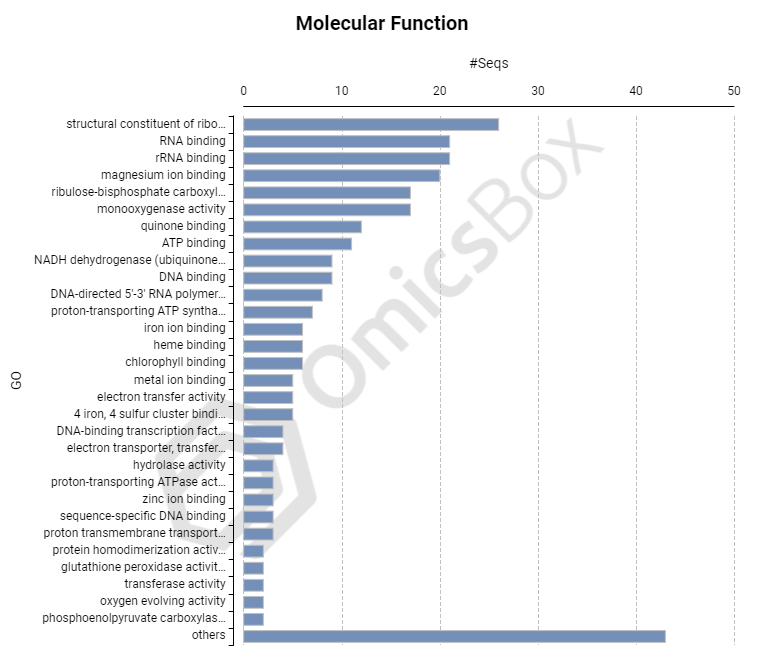
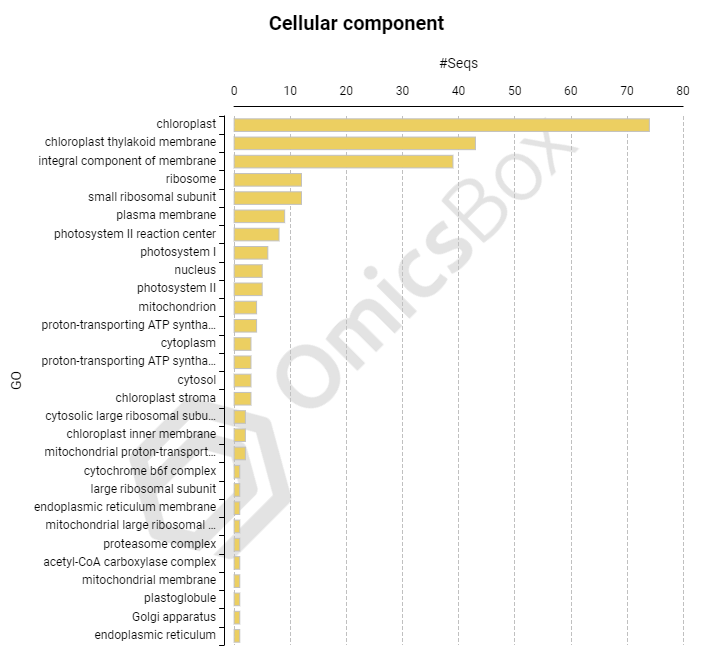
Coding Information
| Acc No. | Species Name | Description | Length | GO IDs | GO Names | Enzyme Names | InterPro GO Names | Peptide Information | BLAST | Nucleotide Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| KP172048.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1337 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX356060.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 212 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| KC584892.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 685 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX356058.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 212 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| GU344680.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1339 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JN895681.1 | Mentha aquatica | maturase K | 745 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| LX078737.1 | Mentha aquatica | linalool synthase | 1821 | P:GO:0016099; P:GO:0016102; P:GO:0033383; F:GO:0000287; F:GO:0010333; F:GO:0030145; F:GO:0034007; F:GO:0034008; C:GO:0009507 | P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; P:geranyl diphosphate metabolic process; F:magnesium ion binding; F:terpene synthase activity; F:manganese ion binding; F:S-linalool synthase activity; F:R-linalool synthase activity; C:chloroplast | S-linalool synthase; R-linalool synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| HM850796.1 | Mentha aquatica | maturase K | 813 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| LX078822.1 | Mentha aquatica | linalool synthase | 1821 | P:GO:0016099; P:GO:0016102; P:GO:0033383; F:GO:0000287; F:GO:0010333; F:GO:0030145; F:GO:0034007; F:GO:0034008; C:GO:0009507 | P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; P:geranyl diphosphate metabolic process; F:magnesium ion binding; F:terpene synthase activity; F:manganese ion binding; F:S-linalool synthase activity; F:R-linalool synthase activity; C:chloroplast | S-linalool synthase; R-linalool synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| MG224103.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 517 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| FR720532.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 551 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KP172053.1 | Mentha aquatica | maturase K | 787 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MH245208.1 | Mentha aquatica | myb-related protein 308-like | 783 | P:GO:0031327; P:GO:2000762; F:GO:0003677 | P:negative regulation of cellular biosynthetic process; P:regulation of phenylpropanoid metabolic process; F:DNA binding | no GO terms | view details | |||
| KX879092.1 | Mentha aquatica | maturase K | 860 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| FR719059.1 | Mentha aquatica | maturase K | 810 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MH208306.2 | Mentha aquatica | cinnamate 4-hydroxylase | 1518 | P:GO:0016114; F:GO:0005506; F:GO:0016710; F:GO:0016712; F:GO:0020037 | P:terpenoid biosynthetic process; F:iron ion binding; F:trans-cinnamate 4-monooxygenase activity; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:heme binding | Trans-cinnamate 4-monooxygenase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no IPS match | view details | ||
| HM850170.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 684 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX879090.1 | Mentha aquatica | maturase K | 858 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| FR719058.1 | Mentha aquatica | maturase K | 810 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KX879091.1 | Mentha aquatica | 859 | P:mRNA processing; C:chloroplast | view details | ||||||
| AY083653.1 | Mentha aquatica | linalool synthase | 1821 | P:GO:0016099; P:GO:0016102; P:GO:0033383; F:GO:0000287; F:GO:0010333; F:GO:0030145; F:GO:0034007; F:GO:0034008; C:GO:0009507 | P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; P:geranyl diphosphate metabolic process; F:magnesium ion binding; F:terpene synthase activity; F:manganese ion binding; F:S-linalool synthase activity; F:R-linalool synthase activity; C:chloroplast | S-linalool synthase; R-linalool synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| KX356057.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 212 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| MG893900.1 | Mentha aquatica | hydroxyphenylpyruvate reductase | 942 | F:GO:0047995; F:GO:0051287; F:GO:0102742 | F:hydroxyphenylpyruvate reductase activity; F:NAD binding; F:R(+)-3,4-dihydroxyphenyllactate:NADP+ oxidoreductase activity | Hydroxyphenylpyruvate reductase | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| LX078682.1 | Mentha aquatica | linalool synthase | 1821 | P:GO:0016099; P:GO:0016102; P:GO:0033383; F:GO:0000287; F:GO:0010333; F:GO:0030145; F:GO:0034007; F:GO:0034008; C:GO:0009507 | P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; P:geranyl diphosphate metabolic process; F:magnesium ion binding; F:terpene synthase activity; F:manganese ion binding; F:S-linalool synthase activity; F:R-linalool synthase activity; C:chloroplast | S-linalool synthase; R-linalool synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| KX356059.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 212 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| KX879089.1 | Mentha aquatica | maturase K | 861 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| KC584934.1 | Mentha aquatica | maturase K | 818 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| LX078802.1 | Mentha aquatica | linalool synthase | 1821 | P:GO:0016099; P:GO:0016102; P:GO:0033383; F:GO:0000287; F:GO:0010333; F:GO:0030145; F:GO:0034007; F:GO:0034008; C:GO:0009507 | P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; P:geranyl diphosphate metabolic process; F:magnesium ion binding; F:terpene synthase activity; F:manganese ion binding; F:S-linalool synthase activity; F:R-linalool synthase activity; C:chloroplast | S-linalool synthase; R-linalool synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| KY249928.1 | Mentha aquatica | terpene synthase 6 | 256 | P:GO:0009414; P:GO:0016102; P:GO:1901937; F:GO:0000287; F:GO:0042803; F:GO:0080016; F:GO:0080017 | P:response to water deprivation; P:diterpenoid biosynthetic process; P:beta-caryophyllene biosynthetic process; F:magnesium ion binding; F:protein homodimerization activity; F:(-)-E-beta-caryophyllene synthase activity; F:alpha-humulene synthase activity | (-)-beta-caryophyllene synthase; Alpha-humulene synthase | F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| FR720533.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 551 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| GQ241339.1 | Mentha aquatica | glyceraldehyde-3-phosphate dehydrogenase | 1373 | P:GO:0006006; P:GO:0006096; F:GO:0004365; F:GO:0050661; F:GO:0051287; C:GO:0005829; C:GO:0009536 | P:glucose metabolic process; P:glycolytic process; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity; F:NADP binding; F:NAD binding; C:cytosol; C:plastid | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating); Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | view details | ||
| KF735672.1 | Mentha aquatica | Regulator of rDNA transcription protein 15 | 669 | no GO terms | view details | |||||
| MH748575.1 | Mentha aquatica | limonene synthase | 2687 | P:GO:0016102; F:GO:0000287; F:GO:0010333 | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity | Carbon-oxygen lyases | no GO terms | view details | ||
| KM051110.1 | Mentha aquatica | limonene hydroxylase | 930 | P:GO:0009625; P:GO:0009751; P:GO:0009753; P:GO:0010225; P:GO:0016114; F:GO:0005506; F:GO:0018674; F:GO:0018675; F:GO:0020037; F:GO:0052741; C:GO:0005789; C:GO:0016021 | P:response to insect; P:response to salicylic acid; P:response to jasmonic acid; P:response to UV-C; P:terpenoid biosynthetic process; F:iron ion binding; F:(S)-limonene 3-monooxygenase activity; F:(S)-limonene 6-monooxygenase activity; F:heme binding; F:(R)-limonene 6-monooxygenase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | (S)-limonene 6-monooxygenase; (R)-limonene 6-monooxygenase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); (S)-limonene 3-monooxygenase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| MZ190987.1 | Mentha aquatica | hypothetical protein BVRB_017900 | 498 | no GO terms | view details | |||||
| KM051109.1 | Mentha aquatica | limonene hydroxylase | 938 | P:GO:0009625; P:GO:0009751; P:GO:0009753; P:GO:0010225; P:GO:0016114; F:GO:0005506; F:GO:0018674; F:GO:0018675; F:GO:0020037; F:GO:0052741; C:GO:0005789; C:GO:0016021 | P:response to insect; P:response to salicylic acid; P:response to jasmonic acid; P:response to UV-C; P:terpenoid biosynthetic process; F:iron ion binding; F:(S)-limonene 3-monooxygenase activity; F:(S)-limonene 6-monooxygenase activity; F:heme binding; F:(R)-limonene 6-monooxygenase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | (S)-limonene 6-monooxygenase; (R)-limonene 6-monooxygenase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); (S)-limonene 3-monooxygenase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| KZ356059.1 | Mentha aquatica | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 201 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| KI879089.1 | Mentha aquatica | maturase K | 851 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details |
lncRNA Information
Gene Information
| Gene ID | Species Name | Symbol | Aliases | Description | Start position | End position | Orientation | Genomic Context |
|---|
miRNA Information
| Acc. No. | Species Name | Sequence length | Total pre-miRNA | Sequence |
|---|---|---|---|---|
| KF735670.1 | Mentha aquatica | 602 | 7 Details |
|
| KF735673.1 | Mentha aquatica | 502 | 5 Details |
RNA-Seq Information
| Experiment Acc. | Species Name | SRA Study | SRA Run | Treatment | Development Stage | Tissue | Data Accesss |
|---|---|---|---|---|---|---|---|
| ERX5310029 | Mentha aquatica | ERP127839 | ERR5529568 | ||||
| ERX5550976 | Mentha aquatica | ERX5550976 | ERR5909849 | ||||
| ERX5550980 | Mentha aquatica | ERP129004 | ERR5909853 | ||||
| ERX5550979 | Mentha aquatica | ERP129004 | ERR5909852 | ||||
| ERX5550978 | Mentha aquatica | ERP129004 | ERR5909851 | ||||
| ERX5550977 | Mentha aquatica | ERP129004 | ERR5909850 | ||||
| ERX5550975 | Mentha aquatica | ERP129004 | ERR5909848 | ||||
| ERX5550974 | Mentha aquatica | ERP129004 | ERR5909847 | ||||
| ERX5550973 | Mentha aquatica | ERP129004 | ERR5909846 | ||||
| SRX533475 | Mentha aquatica | SRP041687 | SRR1271575 | ||||
| SRX533474 | Mentha aquatica | SRP041687 | SRR1271574 |
Other non-coding Sequence Information
| Acc. No | Species Name | Length | GC content | Status | Sequence |
|---|---|---|---|---|---|
| KR611529.1 | Mentha aquatica | 588 | 64.35% | Non-coding | |
| KF735671.1 | Mentha aquatica | 606 | 65.40% | Non-coding |
* If data is not displayed please access the respective tab