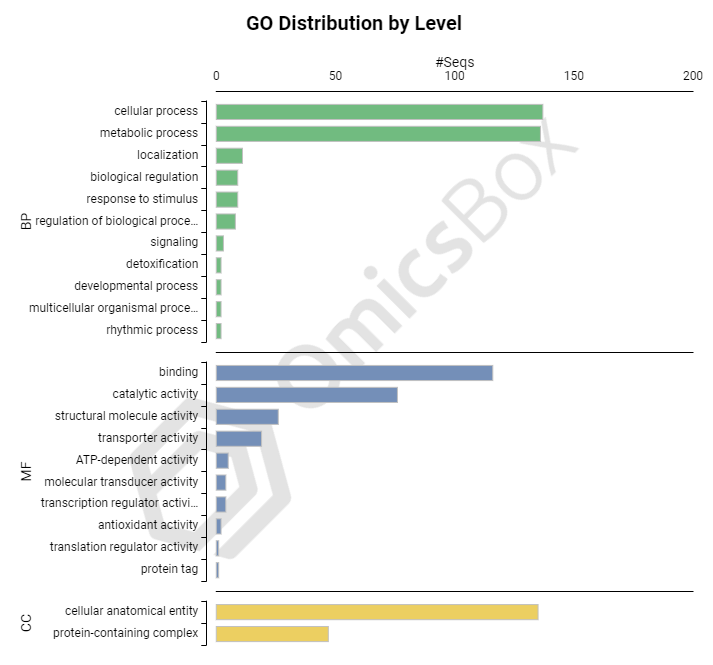
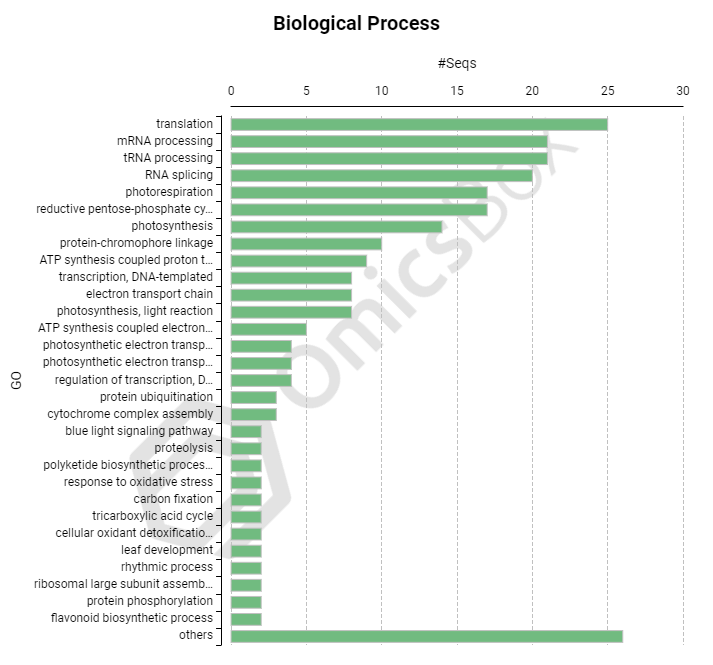
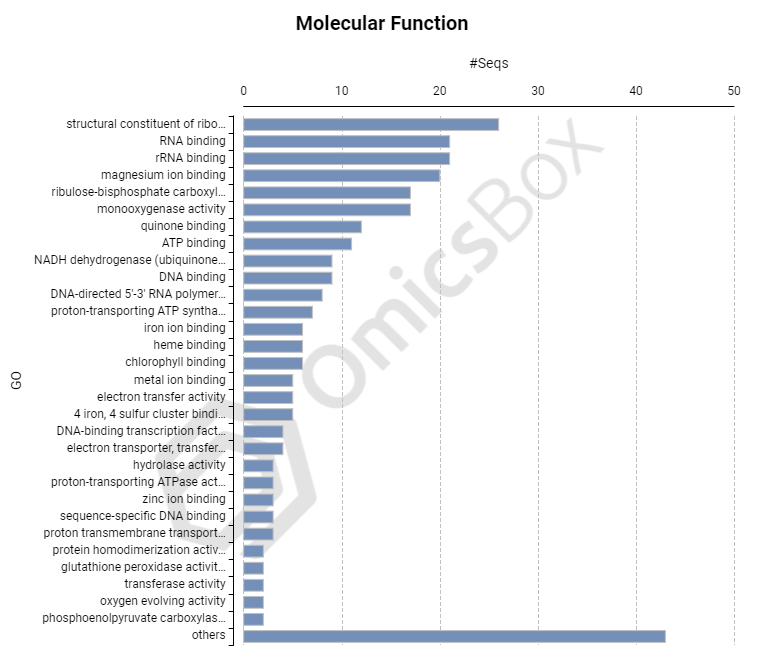
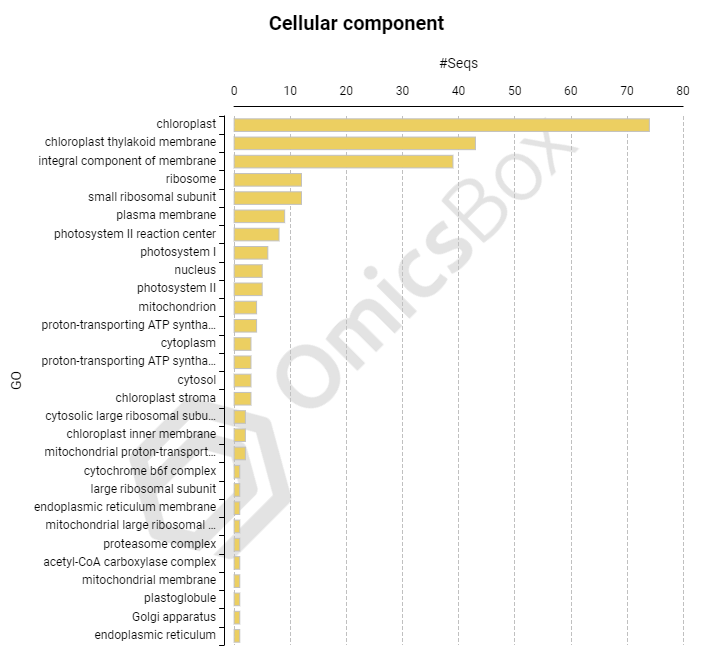
Coding Information
| Acc No. | Species Name | Description | Length | GO IDs | GO Names | Enzyme Names | InterPro GO Names | Peptide Information | BLAST | Nucleotide Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| KX356045.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 217 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| KX879076.1 | Mentha arvensis | maturase K | 842 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130390.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 184 | no GO terms | view details | |||||
| CV130371.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 171 | no GO terms | view details | |||||
| CV130386.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 191 | no GO terms | view details | |||||
| CV130423.1 | Mentha arvensis | transposase | 419 | P:GO:0006313; F:GO:0003677; F:GO:0004803 | P:transposition, DNA-mediated; F:DNA binding; F:transposase activity | no GO terms | view details | |||
| CV130392.1 | Mentha arvensis | uncharacterized protein LOC111475969 | 412 | no IPS match | view details | |||||
| CV130395.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 295 | no GO terms | view details | |||||
| CV130407.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 477 | no GO terms | view details | |||||
| KX879080.1 | Mentha arvensis | maturase K | 809 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MK526182.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 549 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| CV130377.1 | Mentha arvensis | 283 | no GO terms | view details | ||||||
| MG223982.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| LC126639.1 | Mentha arvensis | maturase K | 820 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| JZ468955.1 | Mentha arvensis | ribulose bisphosphate carboxylase small chain 1, chloroplastic-like | 218 | P:GO:0009853; P:GO:0015977; P:GO:0015979; C:GO:0009536 | P:photorespiration; P:carbon fixation; P:photosynthesis; C:plastid | no GO terms | view details | |||
| MF687479.1 | Mentha arvensis | ribosomal protein S14 | 303 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0005840; C:GO:0009507 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:ribosome; C:chloroplast | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| CV130348.1 | Mentha arvensis | putative senescence-associated protein | 288 | no GO terms | view details | |||||
| MG219672.1 | Mentha arvensis | Hypothetical predicted protein | 420 | no IPS match | view details | |||||
| KU174203.1 | Mentha arvensis | menthol dehydrogenase | 762 | F:GO:0047504 | F:(-)-menthol dehydrogenase activity | (-)-menthol dehydrogenase | F:oxidoreductase activity | view details | ||
| MG222273.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MG225072.1 | Mentha arvensis | maturase K | 778 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| JZ468977.1 | Mentha arvensis | nucleoid-associated protein At4g30620, chloroplastic-like | 246 | F:GO:0003677 | F:DNA binding | F:DNA binding | view details | |||
| CV130382.1 | Mentha arvensis | hypothetical protein FEM48_ZijujUnG0010300 | 261 | no GO terms | view details | |||||
| CV130354.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 171 | no GO terms | view details | |||||
| JZ468946.1 | Mentha arvensis | syntaxin-43-like isoform X2 | 316 | P:GO:0006886; P:GO:0006906; P:GO:0048278; F:GO:0000149; F:GO:0005484; C:GO:0012505; C:GO:0016021; C:GO:0031201 | P:intracellular protein transport; P:vesicle fusion; P:vesicle docking; F:SNARE binding; F:SNAP receptor activity; C:endomembrane system; C:integral component of membrane; C:SNARE complex | P:vesicle-mediated transport; C:membrane | view details | |||
| JZ468983.1 | Mentha arvensis | putative receptor-like protein kinase At4g00960 isoform X1 | 140 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | no IPS match | view details | ||
| CV130357.1 | Mentha arvensis | putative senescence-associated protein | 447 | no GO terms | view details | |||||
| MG222509.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| CV130359.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 294 | no GO terms | view details | |||||
| CV130351.1 | Mentha arvensis | uncharacterized protein LOC111475969 | 289 | no GO terms | view details | |||||
| JN893654.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 458 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MG224998.1 | Mentha arvensis | maturase K | 764 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130370.1 | Mentha arvensis | cytochrome P450-like TBP protein | 197 | no GO terms | view details | |||||
| JZ468948.1 | Mentha arvensis | ethylene-responsive transcription factor RAP2-13-like | 83 | P:GO:0006355; P:GO:0006952; P:GO:0009408; P:GO:0009409; P:GO:0009414; P:GO:0009416; P:GO:0009611; P:GO:0009736; P:GO:0009873; P:GO:0045595; P:GO:0071472; F:GO:0000976; F:GO:0003700; F:GO:0005515; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:defense response; P:response to heat; P:response to cold; P:response to water deprivation; P:response to light stimulus; P:response to wounding; P:cytokinin-activated signaling pathway; P:ethylene-activated signaling pathway; P:regulation of cell differentiation; P:cellular response to salt stress; F:transcription cis-regulatory region binding; F:DNA-binding transcription factor activity; F:protein binding; C:nucleus | no IPS match | view details | |||
| CV130353.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 175 | no GO terms | view details | |||||
| JN896123.1 | Mentha arvensis | maturase K | 910 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507; C:GO:0016021 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast; C:integral component of membrane | P:mRNA processing; C:chloroplast | view details | |||
| JZ468952.1 | Mentha arvensis | hypothetical protein SASPL_130169 | 307 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | no IPS match | view details | |||
| KX356048.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 217 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| CV130374.1 | Mentha arvensis | putative senescence-associated protein | 448 | no GO terms | view details | |||||
| FJ999995.1 | Mentha arvensis | cis-muuroladiene synthase-like | 1665 | P:GO:0006952; P:GO:0016102; P:GO:0051762; F:GO:0000287; F:GO:0010334; C:GO:0005737 | P:defense response; P:diterpenoid biosynthetic process; P:sesquiterpene biosynthetic process; F:magnesium ion binding; F:sesquiterpene synthase activity; C:cytoplasm | Carbon-oxygen lyases | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| CV130363.1 | Mentha arvensis | hypothetical protein FEM48_ZijujUnG0010300 | 203 | no GO terms | view details | |||||
| LC126636.1 | Mentha arvensis | maturase K | 820 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| HM359217.1 | Mentha arvensis | unnamed protein product | 291 | F:GO:0005524; C:GO:0005856 | F:ATP binding; C:cytoskeleton | no GO terms | view details | |||
| JZ468957.1 | Mentha arvensis | putative receptor-like protein kinase At4g00960 isoform X1 | 137 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | no IPS match | view details | ||
| HQ590183.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 587 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JZ468963.1 | Mentha arvensis | chlorsulfuron/imidazolinone resistant 1 | 1048 | P:GO:0009097; P:GO:0009099; P:GO:0009635; F:GO:0000287; F:GO:0003984; F:GO:0030976; F:GO:0050660; C:GO:0005948 | P:isoleucine biosynthetic process; P:valine biosynthetic process; P:response to herbicide; F:magnesium ion binding; F:acetolactate synthase activity; F:thiamine pyrophosphate binding; F:flavin adenine dinucleotide binding; C:acetolactate synthase complex | Acetolactate synthase | F:magnesium ion binding; F:catalytic activity; F:thiamine pyrophosphate binding | view details | ||
| MN401228.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 870 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KU174202.1 | Mentha arvensis | menthol dehydrogenase | 942 | F:GO:0000166; F:GO:0047501; F:GO:0047504 | F:nucleotide binding; F:(+)-neomenthol dehydrogenase activity; F:(-)-menthol dehydrogenase activity | (+)-neomenthol dehydrogenase; (-)-menthol dehydrogenase | F:oxidoreductase activity | view details | ||
| KX356046.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 217 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| LC126637.1 | Mentha arvensis | maturase K | 820 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| KX356044.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 217 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| KX879078.1 | Mentha arvensis | maturase K | 842 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130362.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 295 | no GO terms | view details | |||||
| CV130367.1 | Mentha arvensis | putative senescence-associated protein | 452 | no GO terms | view details | |||||
| KC571806.1 | Mentha arvensis | maturase K | 644 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MZ190986.1 | Mentha arvensis | hypothetical protein BVRB_017900 | 444 | no GO terms | view details | |||||
| CV130394.1 | Mentha arvensis | putative senescence-associated protein | 138 | no GO terms | view details | |||||
| CV130364.1 | Mentha arvensis | putative senescence-associated protein | 407 | no GO terms | view details | |||||
| CV130387.1 | Mentha arvensis | senescence-associated protein | 419 | no GO terms | view details | |||||
| MG222778.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| CV130385.1 | Mentha arvensis | uncharacterized protein LOC111475745, partial | 315 | no GO terms | view details | |||||
| LC126640.1 | Mentha arvensis | maturase K | 820 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MG225043.1 | Mentha arvensis | maturase K | 778 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130376.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 292 | no GO terms | view details | |||||
| CV130393.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 170 | no GO terms | view details | |||||
| CV130388.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 423 | no GO terms | view details | |||||
| KX356047.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 217 | P:GO:0009853; P:GO:0019253; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | no IPS match | view details | ||
| CV130369.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 293 | no GO terms | view details | |||||
| LC122466.1 | Mentha arvensis | hypothetical protein SASPL_141406 | 365 | no GO terms | view details | |||||
| KP172052.1 | Mentha arvensis | maturase K | 776 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| CV130358.1 | Mentha arvensis | putative senescence-associated protein | 448 | no GO terms | view details | |||||
| KP172047.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 576 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| CV130356.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 295 | no GO terms | view details | |||||
| KC571787.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 556 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JF289043.1 | Mentha arvensis | Ycf1 | 5105 | P:GO:0015031; C:GO:0009706; C:GO:0016021 | P:protein transport; C:chloroplast inner membrane; C:integral component of membrane | no GO terms | view details | |||
| CV130368.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 293 | no GO terms | view details | |||||
| EF546776.1 | Mentha arvensis | limonene hydroxylase | 1503 | P:GO:0009751; P:GO:0009753; P:GO:0010225; P:GO:0016114; F:GO:0005506; F:GO:0018674; F:GO:0018675; F:GO:0020037; F:GO:0052741; C:GO:0005789; C:GO:0016021 | P:response to salicylic acid; P:response to jasmonic acid; P:response to UV-C; P:terpenoid biosynthetic process; F:iron ion binding; F:(S)-limonene 3-monooxygenase activity; F:(S)-limonene 6-monooxygenase activity; F:heme binding; F:(R)-limonene 6-monooxygenase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | (S)-limonene 6-monooxygenase; (R)-limonene 6-monooxygenase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); (S)-limonene 3-monooxygenase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| CV130360.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 293 | no GO terms | view details | |||||
| MG222963.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| LC126638.1 | Mentha arvensis | maturase K | 820 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| KR072659.1 | Mentha arvensis | ribosomal protein S11 | 417 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0005840; C:GO:0009507 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:ribosome; C:chloroplast | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| KC571788.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 518 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| CV130375.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 195 | no GO terms | view details | |||||
| JZ468978.1 | Mentha arvensis | putative receptor-like protein kinase At4g00960 isoform X2 | 127 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | no IPS match | view details | ||
| KU174204.1 | Mentha arvensis | menthol dehydrogenase | 468 | P:GO:0031525; P:GO:0042214; F:GO:0047504; F:GO:0052581; F:GO:0070402; C:GO:0005737 | P:menthol biosynthetic process; P:terpene metabolic process; F:(-)-menthol dehydrogenase activity; F:(-)-isopiperitenone reductase activity; F:NADPH binding; C:cytoplasm | (-)-menthol dehydrogenase; (-)-isopiperitenone reductase | F:oxidoreductase activity | view details | ||
| MK520309.1 | Mentha arvensis | maturase K | 782 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MG224956.1 | Mentha arvensis | maturase K | 777 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| JZ468951.1 | Mentha arvensis | ethylene-responsive transcription factor RAP2-4-like | 910 | P:GO:0006355; P:GO:0006952; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:defense response; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| KC571805.1 | Mentha arvensis | maturase K | 743 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130373.1 | Mentha arvensis | senescence-associated protein | 447 | no IPS match | view details | |||||
| KX879077.1 | Mentha arvensis | maturase K | 848 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130352.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 188 | no GO terms | view details | |||||
| MN380710.1 | Mentha arvensis | maturase K | 555 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| KJ841415.1 | Mentha arvensis | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 462 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX879079.1 | Mentha arvensis | maturase K | 839 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| CV130349.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 170 | no GO terms | view details | |||||
| CV130344.1 | Mentha arvensis | Regulator of rDNA transcription protein 15 | 170 | no GO terms | view details | |||||
| JZ468979.1 | Mentha arvensis | DNA-(apurinic or apyrimidinic site) lyase 2 | 148 | P:GO:0006284; P:GO:0006629; P:GO:0080111; P:GO:0090305; P:GO:0098506; F:GO:0003677; F:GO:0003906; F:GO:0004519; F:GO:0008081; F:GO:0008270; F:GO:0008311; F:GO:0016829; F:GO:0046403; C:GO:0005634; C:GO:0005746 | P:base-excision repair; P:lipid metabolic process; P:DNA demethylation; P:nucleic acid phosphodiester bond hydrolysis; P:polynucleotide 3 dephosphorylation; F:DNA binding; F:DNA-(apurinic or apyrimidinic site) endonuclease activity; F:endonuclease activity; F:phosphoric diester hydrolase activity; F:zinc ion binding; F:double-stranded DNA 3-5 exodeoxyribonuclease activity; F:lyase activity; F:polynucleotide 3-phosphatase activity; C:nucleus; C:mitochondrial respirasome | Acting on ester bonds; Acting on ester bonds; Lyases; Acting on ester bonds; Polynucleotide 3-phosphatase | P:DNA repair; F:nuclease activity | view details | ||
| EU108705.1 | Mentha arvensis | menthofuran synthase | 1559 | P:GO:0016114; F:GO:0005506; F:GO:0016712; F:GO:0020037; F:GO:0052582; C:GO:0016021; C:GO:0043231 | P:terpenoid biosynthetic process; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:heme binding; F:(+)-menthofuran synthase activity; C:integral component of membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); (+)-menthofuran synthase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JX555963.1 | Mentha arvensis | limonene synthase | 2993 | P:GO:0016102; F:GO:0000287; F:GO:0010333 | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity | Carbon-oxygen lyases | F:terpene synthase activity; F:lyase activity | view details | ||
| EU108706.1 | Mentha arvensis | menthofuran synthase | 1559 | P:GO:0016114; F:GO:0005506; F:GO:0016712; F:GO:0020037; F:GO:0052582; C:GO:0016021; C:GO:0043231 | P:terpenoid biosynthetic process; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:heme binding; F:(+)-menthofuran synthase activity; C:integral component of membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); (+)-menthofuran synthase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| KX267734.1 | Mentha arvensis | ethylene-responsive transcription factor RAP2-4-like | 1477 | P:GO:0006355; P:GO:0006952; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:defense response; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| CV130417.1 | Mentha arvensis | pathogenesis-related protein STH-21-like | 438 | P:GO:0006952; P:GO:0009607; P:GO:0009738; P:GO:0043086; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:defense response; P:response to biotic stimulus; P:abscisic acid-activated signaling pathway; P:negative regulation of catalytic activity; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | no IPS match | view details | |||
| JZ468969.1 | Mentha arvensis | fructose-1,6-bisphosphatase, cytosolic | 917 | P:GO:0005975; P:GO:0016311; F:GO:0042132; F:GO:0046872 | P:carbohydrate metabolic process; P:dephosphorylation; F:fructose 1,6-bisphosphate 1-phosphatase activity; F:metal ion binding | Sugar-phosphatase; Fructose-bisphosphatase | P:carbohydrate metabolic process; F:phosphatase activity; F:fructose 1,6-bisphosphate 1-phosphatase activity | view details | ||
| JZ468945.1 | Mentha arvensis | ---NA--- | 779 | no IPS match | view details | |||||
| CV130412.1 | Mentha arvensis | 483 | no IPS match | view details | ||||||
| JZ468958.1 | Mentha arvensis | beta-carotene isomerase D27, chloroplastic-like | 922 | F:GO:0005506; F:GO:0016853 | F:iron ion binding; F:isomerase activity | Isomerases | F:iron ion binding | view details | ||
| KC591661.1 | Mentha arvensis | hypothetical protein SASPL_127185 | 734 | no GO terms | view details | |||||
| JZ468966.1 | Mentha arvensis | 843 | no GO terms | view details | ||||||
| JZ468960.1 | Mentha arvensis | 1118 | no GO terms | view details | ||||||
| EU108708.1 | Mentha arvensis | 1609 | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||||||
| EU108707.1 | Mentha arvensis | menthofuran synthase | 1609 | P:GO:0016114; F:GO:0005506; F:GO:0016712; F:GO:0020037; F:GO:0052582; C:GO:0016021; C:GO:0043231 | P:terpenoid biosynthetic process; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:heme binding; F:(+)-menthofuran synthase activity; C:integral component of membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); (+)-menthofuran synthase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| LC050897.1 | Mentha arvensis | RNA polymerase beta subunit | 603 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0032549; C:GO:0000428; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding; C:DNA-directed RNA polymerase complex; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding | view details |
lncRNA Information
Gene Information
| Gene ID | Species Name | Symbol | Aliases | Description | Start position | End position | Orientation | Genomic Context |
|---|
miRNA Information
| Acc. No. | Species Name | Sequence length | Total pre-miRNA | Sequence |
|---|---|---|---|---|
| JZ979502.1 | Mentha arvensis | 793 | 6 Details |
|
| CV130347.1 | Mentha arvensis | 195 | 1 Details |
|
| CV130346.1 | Mentha arvensis | 169 | 1 Details |
|
| CV130365.1 | Mentha arvensis | 195 | 1 Details |
RNA-Seq Information
| Experiment Acc. | Species Name | SRA Study | SRA Run | Treatment | Development Stage | Tissue | Data Accesss |
|---|---|---|---|---|---|---|---|
| SRX8861008 | Mentha arvensis | ?SRP274606 | SRR12361980 | ||||
| SRX6096639 | Mentha arvensis | SRP201939 | SRR9329904 | 8 day waterlogging | Root | ||
| SRX6096638 | Mentha arvensis | ?SRP201939 | SRR9329905 | Control water logging | Root | ||
| SRX6096637 | Mentha arvensis | ?SRP201939 | SRR9329906 | 8 day waterlogging | Root | ||
| SRX6096636 | Mentha arvensis | SRP201939 | SRR9329907 | Control water logging | Root | ||
| SRX2507777 | Mentha arvensis | SRP097317 | SRR5192063 | early | leaf | ||
| SRX2507776 | Mentha arvensis | SRP097317 | SRR5192062 | late | |||
| SRX1842797 | Mentha arvensis | SRP076515 | SRR3664035 | ||||
| SRX1600048 | Mentha arvensis | SRP070802 | SRR3187004 | trichome rich | |||
| SRX533469 | Mentha arvensis | SRP041687 | SRR1271560 | ||||
| SRX533468 | Mentha arvensis | SRP041687 | SRR1271559 |
Other non-coding Sequence Information
* If data is not displayed please access the respective tab