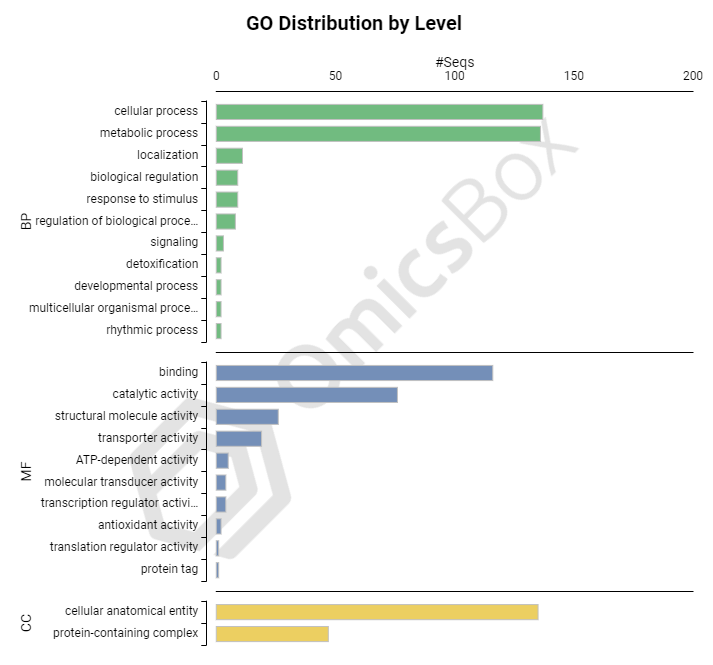
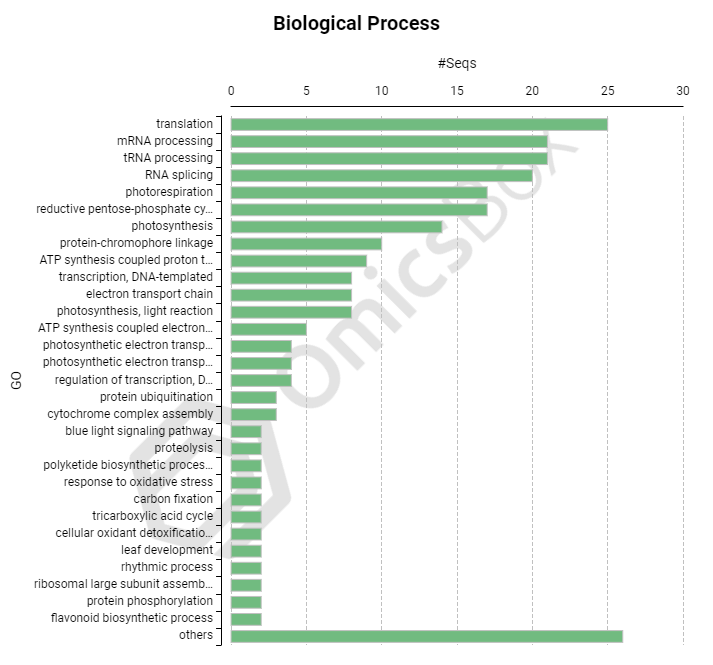
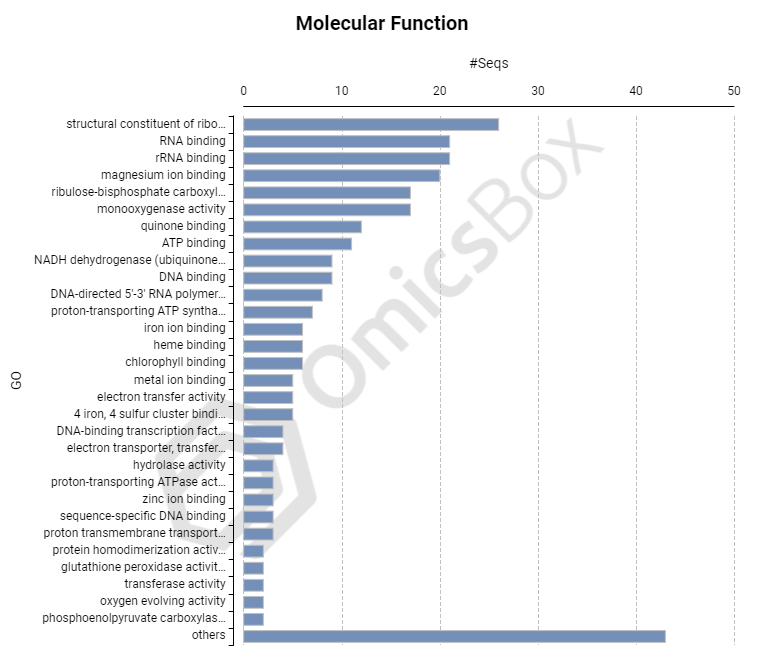
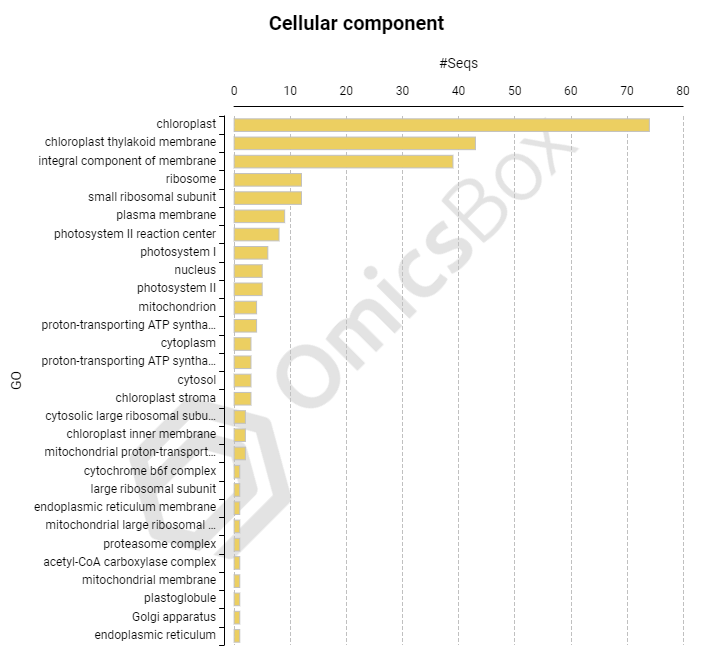
Coding Information
| Acc No. | Species Name | Description | Length | GO IDs | GO Names | Enzyme Names | InterPro GO Names | Peptide Information | BLAST | Nucleotide Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| MF379677.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MF468156.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MF379678.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| FR720543.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 551 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF468149.2 | Ocimum tenuiflorum | maturase K | 866 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MF468174.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442493.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 983 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| KC571804.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 472 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442482.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1007 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF468148.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| HM990148.1 | Ocimum tenuiflorum | 4-coumarate--CoA ligase 2-like | 1704 | P:GO:0009698; F:GO:0016207; F:GO:0106290 | P:phenylpropanoid metabolic process; F:4-coumarate-CoA ligase activity; F:trans-cinnamate-CoA ligase activity | 4-coumarate--CoA ligase | no GO terms | view details | ||
| MK442484.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1007 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| HM990149.1 | Ocimum tenuiflorum | 1518 | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||||||
| HM990153.1 | Ocimum tenuiflorum | caffeic acid O-methyltransferase | 1086 | P:GO:0009809; P:GO:0032259; F:GO:0046983; F:GO:0047763 | P:lignin biosynthetic process; P:methylation; F:protein dimerization activity; F:caffeate O-methyltransferase activity | Caffeate O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MK442481.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1005 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| EU622046.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1070 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF468180.2 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MN660047.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 740 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| EU622042.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1043 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MK442478.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1007 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| KX898241.1 | Ocimum tenuiflorum | maturase K | 782 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MF379681.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| HO001941.1 | Ocimum tenuiflorum | cytochrome P450 81Q32 | 674 | P:GO:0051762; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:sesquiterpene biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| OL537014.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 553 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF468177.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK090294.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 559 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KC571803.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 555 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| LC461782.1 | Ocimum tenuiflorum | maturase K | 730 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| JX465682.1 | Ocimum tenuiflorum | maturase K | 798 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| JF357829.1 | Ocimum tenuiflorum | maturase K | 797 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MK442490.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 984 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF326417.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 511 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442489.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1001 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MK442492.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1007 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF379679.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MK442477.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 994 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| HO001936.1 | Ocimum tenuiflorum | 4-coumarate--CoA ligase 2-like | 713 | P:GO:0009698; F:GO:0016207; F:GO:0106290 | P:phenylpropanoid metabolic process; F:4-coumarate-CoA ligase activity; F:trans-cinnamate-CoA ligase activity | 4-coumarate--CoA ligase | no GO terms | view details | ||
| EU622048.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1044 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MN542659.1 | Ocimum tenuiflorum | hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase | 1278 | F:GO:0050266 | F:rosmarinate synthase activity | Rosmarinate synthase | no GO terms | view details | ||
| MK442488.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1007 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF468152.2 | Ocimum tenuiflorum | maturase K | 865 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MN393294.1 | Ocimum tenuiflorum | probable WRKY transcription factor 53 | 1023 | P:GO:0006355; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding | view details | |||
| MF326416.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 511 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO001930.1 | Ocimum tenuiflorum | cytochrome P450 81Q32 | 659 | P:GO:0051762; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:sesquiterpene biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| KP208911.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 537 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442479.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 947 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| KU856609.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 532 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF468176.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF468154.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MK442486.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 984 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JN114765.1 | Ocimum tenuiflorum | maturase K | 711 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MF326413.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 511 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HM990156.1 | Ocimum tenuiflorum | cytochrome P450 | 1530 | P:GO:0009805; P:GO:0009809; P:GO:0009813; P:GO:0016114; F:GO:0005506; F:GO:0016712; F:GO:0020037; F:GO:0042802; F:GO:0046409; C:GO:0005783; C:GO:0016021 | P:coumarin biosynthetic process; P:lignin biosynthetic process; P:flavonoid biosynthetic process; P:terpenoid biosynthetic process; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:heme binding; F:identical protein binding; F:p-coumarate 3-hydroxylase activity; C:endoplasmic reticulum; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JX465680.1 | Ocimum tenuiflorum | maturase K | 809 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| EU622043.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1030 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF468178.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| HO001931.1 | Ocimum tenuiflorum | cytochrome P450 81Q32 | 128 | P:GO:0051762; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:sesquiterpene biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no IPS match | view details | ||
| MF468155.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MF399413.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MK442491.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1001 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF326412.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 511 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF379676.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MF326411.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 498 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JN114828.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 613 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF468173.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX898240.1 | Ocimum tenuiflorum | maturase K | 773 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MK442480.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1009 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| KC571821.1 | Ocimum tenuiflorum | maturase K | 713 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MW628932.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 517 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MH925775.1 | Ocimum tenuiflorum | (3S,6E)-nerolidol synthase 1-like isoform X1 | 1726 | P:GO:0016114; F:GO:0000287; F:GO:0010333 | P:terpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity | Carbon-oxygen lyases | F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| MF468151.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MH925776.1 | Ocimum tenuiflorum | linalool synthase | 1410 | P:GO:0016099; P:GO:0016102; F:GO:0000287; F:GO:0050437; F:GO:0050551; C:GO:0009507 | P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; F:magnesium ion binding; F:(-)-endo-fenchol synthase activity; F:myrcene synthase activity; C:chloroplast | (-)-endo-fenchol synthase; Myrcene synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| FR719068.1 | Ocimum tenuiflorum | maturase K | 801 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| KU977436.1 | Ocimum tenuiflorum | eugenol synthase | 945 | P:GO:0042855; F:GO:0000166; F:GO:0016491; F:GO:0042803 | P:eugenol biosynthetic process; F:nucleotide binding; F:oxidoreductase activity; F:protein homodimerization activity | Oxidoreductases | F:oxidoreductase activity | view details | ||
| EU622044.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1040 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| KF381105.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 582 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442487.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1007 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| EU622045.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1066 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| EU622049.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 977 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF468175.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF468153.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| MF326415.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 511 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KU977435.1 | Ocimum tenuiflorum | eugenol synthase | 945 | P:GO:0042855; F:GO:0000166; F:GO:0016491; F:GO:0042803 | P:eugenol biosynthetic process; F:nucleotide binding; F:oxidoreductase activity; F:protein homodimerization activity | Oxidoreductases | F:oxidoreductase activity | view details | ||
| KP172063.1 | Ocimum tenuiflorum | maturase K | 726 | P:GO:0006397; P:GO:0008033; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| KX898239.1 | Ocimum tenuiflorum | maturase K | 773 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MF379680.1 | Ocimum tenuiflorum | maturase K | 720 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| MT521743.1 | Ocimum tenuiflorum | phenylalanine ammonia-lyase | 223 | P:GO:0006559; P:GO:0009800; F:GO:0045548; C:GO:0005737 | P:L-phenylalanine catabolic process; P:cinnamic acid biosynthetic process; F:phenylalanine ammonia-lyase activity; C:cytoplasm | Phenylalanine ammonia-lyase; Phenylalanine/tyrosine ammonia-lyase | F:catalytic activity | view details | ||
| MH925777.1 | Ocimum tenuiflorum | 1,8-cineole synthase | 1743 | P:GO:0010597; P:GO:0016099; P:GO:0016102; P:GO:0046248; F:GO:0000287; F:GO:0050550; F:GO:0050551; F:GO:0080015; F:GO:0102313; C:GO:0009507 | P:green leaf volatile biosynthetic process; P:monoterpenoid biosynthetic process; P:diterpenoid biosynthetic process; P:alpha-pinene biosynthetic process; F:magnesium ion binding; F:pinene synthase activity; F:myrcene synthase activity; F:sabinene synthase activity; F:1,8-cineole synthase activity; C:chloroplast | 1,8-cineole synthase; Carbon-oxygen lyases; Myrcene synthase | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| MF468179.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| LC461783.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 552 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442485.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1009 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| MF468150.2 | Ocimum tenuiflorum | maturase K | 863 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| OL690083.1 | Ocimum tenuiflorum | maturase K | 819 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JX465692.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 638 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MF326414.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 511 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MH069806.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 605 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KP172072.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1411 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| EU139474.1 | Ocimum tenuiflorum | polyphenol oxidase, chloroplastic-like | 417 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JX465690.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 637 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| MK442483.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1009 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| HM990151.1 | Ocimum tenuiflorum | cinnamyl alcohol dehydrogenase 1 | 1074 | P:GO:0009809; F:GO:0008270; F:GO:0045551; F:GO:0047924; F:GO:0052747 | P:lignin biosynthetic process; F:zinc ion binding; F:cinnamyl-alcohol dehydrogenase activity; F:geraniol dehydrogenase activity; F:sinapyl alcohol dehydrogenase activity | Cinnamyl-alcohol dehydrogenase; Geraniol dehydrogenase (NADP(+)) | no GO terms | view details | ||
| MF468181.1 | Ocimum tenuiflorum | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 743 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| KX898238.1 | Ocimum tenuiflorum | maturase K | 772 | P:GO:0006397; P:GO:0008033; P:GO:0008380; P:GO:0018108; F:GO:0003723; F:GO:0004715; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; P:peptidyl-tyrosine phosphorylation; F:RNA binding; F:non-membrane spanning protein tyrosine kinase activity; C:chloroplast | Non-specific protein-tyrosine kinase | P:mRNA processing; C:chloroplast | view details | ||
| EU622047.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 1051 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| GU810519.1 | Ocimum tenuiflorum | Protein TAR1 | 330 | no GO terms | view details | |||||
| MK087938.1 | Ocimum tenuiflorum | Hypothetical predicted protein | 451 | no GO terms | view details | |||||
| HO001935.1 | Ocimum tenuiflorum | cytochrome P450 mono-oxygenase | 626 | P:GO:0051762; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:sesquiterpene biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JN114986.1 | Ocimum tenuiflorum | RNA polymerase beta subunit | 438 | P:GO:0006351; F:GO:0003677; F:GO:0003899; C:GO:0000428; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; C:DNA-directed RNA polymerase complex; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity | view details | ||
| FR720482.1 | Ocimum tenuiflorum | RNA polymerase beta subunit | 491 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0032549; C:GO:0000428; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding; C:DNA-directed RNA polymerase complex; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding | view details | ||
| JN114904.1 | Ocimum tenuiflorum | RNA polymerase beta subunit | 362 | P:GO:0006351; F:GO:0003677; F:GO:0003899; F:GO:0032549; C:GO:0000428; C:GO:0009507 | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding; C:DNA-directed RNA polymerase complex; C:chloroplast | DNA-directed RNA polymerase | P:transcription, DNA-templated; F:DNA binding; F:DNA-directed 5-3 RNA polymerase activity; F:ribonucleoside binding | view details | ||
| MW724787.1 | Ocimum tenuiflorum | 151758 | F:ATP binding; F:ATPase | view details | ||||||
| MH883642.1 | Ocimum tenuiflorum | 151744 | F:ATP binding; F:ATPase | view details | ||||||
| HO001942.1 | Ocimum tenuiflorum | probable splicing factor 3A subunit 1 | 707 | P:GO:0045292; F:GO:0003723; C:GO:0005686; C:GO:0071004; C:GO:0071013 | P:mRNA cis splicing, via spliceosome; F:RNA binding; C:U2 snRNP; C:U2-type prespliceosome; C:catalytic step 2 spliceosome | P:mRNA cis splicing, via spliceosome; F:RNA binding | view details | |||
| NC_043873.1 | Ocimum tenuiflorum | 151744 | F:ATP binding; F:ATPase | view details | ||||||
| HO001939.1 | Ocimum tenuiflorum | probable splicing factor 3A subunit 1 | 715 | P:GO:0045292; F:GO:0003723; C:GO:0005681 | P:mRNA cis splicing, via spliceosome; F:RNA binding; C:spliceosomal complex | P:mRNA cis splicing, via spliceosome; F:RNA binding | view details | |||
| HO001932.1 | Ocimum tenuiflorum | probable splicing factor 3A subunit 1 | 706 | P:GO:0045292; F:GO:0003723; C:GO:0005686; C:GO:0071004; C:GO:0071013 | P:mRNA cis splicing, via spliceosome; F:RNA binding; C:U2 snRNP; C:U2-type prespliceosome; C:catalytic step 2 spliceosome | P:mRNA cis splicing, via spliceosome; F:RNA binding | view details | |||
| MN687904.1 | Ocimum tenuiflorum | 152419 | F:ATP binding; F:ATPase | view details | ||||||
| HO001934.1 | Ocimum tenuiflorum | probable splicing factor 3A subunit 1 | 710 | P:GO:0045292; F:GO:0003723; C:GO:0005686; C:GO:0071004; C:GO:0071013 | P:mRNA cis splicing, via spliceosome; F:RNA binding; C:U2 snRNP; C:U2-type prespliceosome; C:catalytic step 2 spliceosome | P:mRNA cis splicing, via spliceosome; F:RNA binding | view details | |||
| MW582310.1 | Ocimum tenuiflorum | eugenol O-methyltransferase | 955 | P:GO:0032259; F:GO:0046983; F:GO:0050630; F:GO:0102719 | P:methylation; F:protein dimerization activity; F:(iso)eugenol O-methyltransferase activity; F:S-adenosyl-L-methionine:eugenol-O-methyltransferase activity | (Iso)eugenol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| MW829604.1 | Ocimum tenuiflorum | 151722 | F:ATP binding; F:ATPase | view details |
lncRNA Information
Gene Information
| Gene ID | Species Name | Symbol | Aliases | Description | Start position | End position | Orientation | Genomic Context |
|---|---|---|---|---|---|---|---|---|
| 40863777 | Ocimum tenuiflorum | rps12 | FPH81_pgp042 | ribosomal protein S12 | 68682 | 68807 | minus | |
| 40863767 | Ocimum tenuiflorum | rpl23 | FPH81_pgp001 | ribosomal protein L23 | 149848 | 150129 | plus | |
| 40863762 | Ocimum tenuiflorum | ndhB | FPH81_pgp004 | NADH-plastoquinone oxidoreductase subunit 2 | 139237 | 141444 | plus | |
| 40863761 | Ocimum tenuiflorum | clpP | FPH81_pgp041 | Clp protease proteolytic subunit | 68923 | 70841 | minus | |
| 40863760 | Ocimum tenuiflorum | rps7 | FPH81_pgp005 | ribosomal protein S7 | 138516 | 138983 | plus | |
| 40863753 | Ocimum tenuiflorum | rps15 | FPH81_pgp007 | ribosomal protein S15 | 120860 | 121132 | minus | |
| 40863752 | Ocimum tenuiflorum | ndhH | FPH81_pgp008 | NADH-plastoquinone oxidoreductase subunit 7 | 119564 | 120745 | minus | |
| 40863751 | Ocimum tenuiflorum | ndhA | FPH81_pgp009 | NADH-plastoquinone oxidoreductase subunit 1 | 117448 | 119562 | minus | |
| 40863750 | Ocimum tenuiflorum | ndhI | FPH81_pgp010 | NADH-plastoquinone oxidoreductase subunit I | 116861 | 117367 | minus | |
| 40863749 | Ocimum tenuiflorum | ndhG | FPH81_pgp011 | NADH-plastoquinone oxidoreductase subunit 6 | 115968 | 116498 | minus | |
| 40863748 | Ocimum tenuiflorum | ndhE | FPH81_pgp012 | NADH-plastoquinone oxidoreductase subunit 4L | 115459 | 115764 | minus | |
| 40863747 | Ocimum tenuiflorum | psaC | FPH81_pgp013 | photosystem I subunit VII | 114958 | 115203 | minus | |
| 40863746 | Ocimum tenuiflorum | ndhD | FPH81_pgp014 | NADH-plastoquinone oxidoreductase subunit 4 | 113338 | 114867 | minus | |
| 40863745 | Ocimum tenuiflorum | ccsA | FPH81_pgp015 | cytochrome cheme attachment protein | 112148 | 113119 | plus | |
| 40863743 | Ocimum tenuiflorum | rpl32 | FPH81_pgp016 | ribosomal protein L32 | 111112 | 111285 | plus | |
| 40863742 | Ocimum tenuiflorum | ndhF | FPH81_pgp017 | NADH-plastoquinone oxidoreductase subunit 5 | 108405 | 110642 | minus | |
| 40863735 | Ocimum tenuiflorum | rps7 | FPH81_pgp019 | ribosomal protein S7 | 95526 | 95993 | minus | |
| 40863734 | Ocimum tenuiflorum | ndhB | FPH81_pgp020 | NADH-plastoquinone oxidoreductase subunit 2 | 93065 | 95272 | minus | |
| 40863729 | Ocimum tenuiflorum | rpl23 | FPH81_pgp023 | ribosomal protein L23 | 84380 | 84661 | minus | |
| 40863728 | Ocimum tenuiflorum | rpl2 | FPH81_pgp024 | ribosomal protein L2 | 82883 | 84361 | minus | |
| 40863727 | Ocimum tenuiflorum | rps19 | FPH81_pgp025 | ribosomal protein S19 | 82540 | 82827 | minus | |
| 40863726 | Ocimum tenuiflorum | rpl22 | FPH81_pgp026 | ribosomal protein L22 | 81985 | 82458 | minus | |
| 40863725 | Ocimum tenuiflorum | rps3 | FPH81_pgp027 | ribosomal protein S3 | 81338 | 82000 | minus | |
| 40863724 | Ocimum tenuiflorum | rpl16 | FPH81_pgp028 | ribosomal protein L16 | 79930 | 80289 | minus | |
| 40863723 | Ocimum tenuiflorum | rpl14 | FPH81_pgp029 | ribosomal protein L14 | 79430 | 79798 | minus | |
| 40863722 | Ocimum tenuiflorum | rps8 | FPH81_pgp030 | ribosomal protein S8 | 78854 | 79258 | minus | |
| 40863721 | Ocimum tenuiflorum | infA | FPH81_pgp031 | translational initiation factor 1 | 78484 | 78717 | minus | |
| 40863720 | Ocimum tenuiflorum | rpl36 | FPH81_pgp032 | ribosomal protein L36 | 78275 | 78388 | minus | |
| 40863719 | Ocimum tenuiflorum | rps11 | FPH81_pgp033 | ribosomal protein S11 | 77749 | 78165 | minus | |
| 40863718 | Ocimum tenuiflorum | rpoA | FPH81_pgp034 | RNA polymerase alpha subunit | 76672 | 77679 | minus | |
| 40863717 | Ocimum tenuiflorum | petD | FPH81_pgp035 | cytochrome b6f complex subunit IV | 75976 | 76500 | plus | |
| 40863716 | Ocimum tenuiflorum | petB | FPH81_pgp036 | cytochrome b6 | 74455 | 75114 | plus | |
| 40863715 | Ocimum tenuiflorum | psbH | FPH81_pgp037 | photosystem II phosphoprotein | 73392 | 73628 | plus | |
| 40863714 | Ocimum tenuiflorum | psbN | FPH81_pgp038 | photosystem II protein N | 73173 | 73304 | minus | |
| 40863713 | Ocimum tenuiflorum | psbT | FPH81_pgp039 | photosystem II protein T | 73006 | 73107 | plus | |
| 40863712 | Ocimum tenuiflorum | psbB | FPH81_pgp040 | photosystem II CP47 chlorophyll apoprotein | 71298 | 72824 | plus | |
| 40863711 | Ocimum tenuiflorum | rps12 | FPH81_pgp043 | ribosomal protein S12 | 68682 | 68807 | minus | |
| 40863711 | Ocimum tenuiflorum | rps12 | FPH81_pgp043 | ribosomal protein S12 | 137669 | 137911 | plus | |
| 40863710 | Ocimum tenuiflorum | rpl20 | FPH81_pgp044 | ribosomal protein L20 | 67572 | 67958 | minus | |
| 40863709 | Ocimum tenuiflorum | rps18 | FPH81_pgp045 | ribosomal protein S18 | 67034 | 67339 | plus | |
| 40863708 | Ocimum tenuiflorum | rpl33 | FPH81_pgp046 | ribosomal protein L33 | 66673 | 66873 | plus | |
| 40863707 | Ocimum tenuiflorum | psaJ | FPH81_pgp047 | photosystem I subunit IX | 66090 | 66224 | plus | |
| 40863703 | Ocimum tenuiflorum | petG | FPH81_pgp048 | cytochrome b6f complex subunit V | 65257 | 65370 | plus | |
| 40863702 | Ocimum tenuiflorum | petL | FPH81_pgp049 | cytochrome b6f complex subunit VI | 64994 | 65089 | plus | |
| 40863701 | Ocimum tenuiflorum | psbE | FPH81_pgp050 | photosystem II cytochrome b559 alpha subunit | 63868 | 64119 | minus | |
| 40863700 | Ocimum tenuiflorum | psbF | FPH81_pgp051 | photosystem II cytochrome b559 beta subunit | 63734 | 63853 | minus | |
| 40863699 | Ocimum tenuiflorum | psbL | FPH81_pgp052 | photosystem II protein L | 63594 | 63710 | minus | |
| 40863698 | Ocimum tenuiflorum | psbJ | FPH81_pgp053 | photosystem II protein J | 63337 | 63459 | minus | |
| 40863697 | Ocimum tenuiflorum | petA | FPH81_pgp054 | cytochromef | 61366 | 62328 | plus | |
| 40863696 | Ocimum tenuiflorum | cemA | FPH81_pgp055 | chloroplast envelope membrane protein | 60485 | 61174 | plus | |
| 40863695 | Ocimum tenuiflorum | ycf4 | FPH81_pgp056 | photosystem I assembly proteinYcf4 | 59336 | 59890 | plus | |
| 40863694 | Ocimum tenuiflorum | psaI | FPH81_pgp057 | photosystem I subunit VIII | 58781 | 58891 | plus | |
| 40863693 | Ocimum tenuiflorum | accD | FPH81_pgp058 | acetyl-CoA carboxylase carboxyltransferase beta subunit | 56445 | 58055 | plus | |
| 40863692 | Ocimum tenuiflorum | rbcL | FPH81_pgp059 | ribulose15-bisphosphatecarboxylaseoxygenaselargesubunit | 54368 | 55822 | plus | |
| 40863691 | Ocimum tenuiflorum | atpB | FPH81_pgp060 | ATP synthase CF1 beta subunit | 52112 | 53608 | minus | |
| 40863690 | Ocimum tenuiflorum | atpE | FPH81_pgp061 | ATP synthase CF1 epsilon subunit | 51714 | 52115 | minus | |
| 40863687 | Ocimum tenuiflorum | ndhC | FPH81_pgp062 | NADH-plastoquinone oxidoreductase subunit 3 | 49333 | 49695 | minus | |
| 40863686 | Ocimum tenuiflorum | ndhK | FPH81_pgp063 | NADH-plastoquinone oxidoreductase subunit K | 48604 | 49281 | minus | |
| 40863685 | Ocimum tenuiflorum | ndhJ | FPH81_pgp064 | NADH-plastoquinone oxidoreductase subunit J | 48016 | 48492 | minus | |
| 40863681 | Ocimum tenuiflorum | rps4 | FPH81_pgp065 | ribosomal protein S4 | 44680 | 45285 | minus | |
| 40863678 | Ocimum tenuiflorum | psaA | FPH81_pgp067 | photosystem I P700 apoprotein A1 | 38458 | 40710 | minus | |
| 40863677 | Ocimum tenuiflorum | psaB | FPH81_pgp068 | photosystem I P700 apoprotein A2 | 36228 | 38432 | minus | |
| 40863676 | Ocimum tenuiflorum | rps14 | FPH81_pgp069 | ribosomal protein S14 | 35803 | 36105 | minus | |
| 40863673 | Ocimum tenuiflorum | psbZ | FPH81_pgp070 | photosystem II protein Z | 34911 | 35099 | plus | |
| 40863671 | Ocimum tenuiflorum | psbC | FPH81_pgp071 | photosystem II CP43 chlorophyll apoprotein | 32828 | 34249 | plus | |
| 40863670 | Ocimum tenuiflorum | psbD | FPH81_pgp072 | photosystem II protein D2 | 31819 | 32880 | plus | |
| 40863664 | Ocimum tenuiflorum | psbM | FPH81_pgp073 | photosystem II protein M | 28841 | 28945 | minus | |
| 40863663 | Ocimum tenuiflorum | petN | FPH81_pgp074 | cytochrome b6f complex subunit VIII | 27731 | 27826 | plus | |
| 40863661 | Ocimum tenuiflorum | rpoB | FPH81_pgp075 | RNA polymerase betasubunit | 23111 | 26323 | minus | |
| 40863660 | Ocimum tenuiflorum | rpoC1 | FPH81_pgp076 | RNA polymerase beta | 20265 | 23084 | minus | |
| 40863659 | Ocimum tenuiflorum | rpoC2 | FPH81_pgp077 | RNA polymerase betasubunit | 15931 | 20100 | minus | |
| 40863658 | Ocimum tenuiflorum | rps2 | FPH81_pgp078 | ribosomal protein S2 | 15004 | 15714 | minus | |
| 40863657 | Ocimum tenuiflorum | atpI | FPH81_pgp079 | ATP synthase CF0 subunitIV | 14036 | 14779 | minus | |
| 40863656 | Ocimum tenuiflorum | atpH | FPH81_pgp080 | ATP synthase CF0 subunitIII | 12772 | 13017 | minus | |
| 40863655 | Ocimum tenuiflorum | atpF | FPH81_pgp081 | ATP synthase CF0 subunitI | 11235 | 12475 | minus | |
| 40863654 | Ocimum tenuiflorum | atpA | FPH81_pgp082 | ATP synthase CF1 alpha subunit | 9611 | 11134 | minus | |
| 40863651 | Ocimum tenuiflorum | psbI | FPH81_pgp083 | photosystem II protein I | 7384 | 7494 | plus | |
| 40863650 | Ocimum tenuiflorum | psbK | FPH81_pgp084 | photosystem II protein K | 6805 | 6990 | plus | |
| 40863648 | Ocimum tenuiflorum | rps16 | FPH81_pgp085 | ribosomal protein S16 | 4768 | 4971 | minus | |
| 40863646 | Ocimum tenuiflorum | matK | FPH81_pgp086 | maturase K | 1980 | 3500 | minus | |
| 40863645 | Ocimum tenuiflorum | psbA | FPH81_pgp087 | photosystem II protein D1 | 381 | 1439 | minus | |
| 40863765 | Ocimum tenuiflorum | ycf2 | FPH81_pgp002 | hypothetical chloroplast RF21 | 142693 | 149514 | minus | |
| 40863764 | Ocimum tenuiflorum | ycf15 | FPH81_pgp003 | hypothetical chloroplast RF15 | 142354 | 142602 | minus | |
| 40863754 | Ocimum tenuiflorum | ycf1 | FPH81_pgp006 | hypothetical chloroplast RF19 | 121567 | 127131 | minus | |
| 40863741 | Ocimum tenuiflorum | ycf1 | FPH81_pgp018 | hypothetical chloroplast RF19 | 107381 | 108511 | plus | |
| 40863732 | Ocimum tenuiflorum | ycf15 | FPH81_pgp021 | hypothetical chloroplast RF15 | 91958 | 92155 | plus | |
| 40863731 | Ocimum tenuiflorum | ycf2 | FPH81_pgp022 | hypothetical chloroplast RF21 | 84995 | 91816 | plus | |
| 40863679 | Ocimum tenuiflorum | ycf3 | FPH81_pgp066 | hypothetical chloroplast RF34 | 41495 | 43444 | minus | |
| 40863774 | Ocimum tenuiflorum | FPH81_pgr002 | FPH81_pgr002 | 23S ribosomal RNA | 129003 | 131812 | minus | |
| 40863773 | Ocimum tenuiflorum | FPH81_pgr003 | FPH81_pgr003 | 4.5S ribosomal RNA | 128802 | 128904 | minus | |
| 40863771 | Ocimum tenuiflorum | FPH81_pgr005 | FPH81_pgr005 | 5S ribosomal RNA | 105963 | 106083 | plus | |
| 40863770 | Ocimum tenuiflorum | FPH81_pgr006 | FPH81_pgr006 | 4.5S ribosomal RNA | 105605 | 105707 | plus | |
| 40863759 | Ocimum tenuiflorum | FPH81_pgr001 | FPH81_pgr001 | 16S ribosomal RNA | 134242 | 135731 | minus | |
| 40863757 | Ocimum tenuiflorum | FPH81_pgr004 | FPH81_pgr004 | 5S ribosomal RNA | 128426 | 128546 | minus | |
| 40863739 | Ocimum tenuiflorum | FPH81_pgr007 | FPH81_pgr007 | 23S ribosomal RNA | 102696 | 105505 | plus | |
| 40863737 | Ocimum tenuiflorum | FPH81_pgr008 | FPH81_pgr008 | 16S ribosomal RNA | 98777 | 100266 | plus | |
| 40863776 | Ocimum tenuiflorum | trnV-GAC | FPH81_pgt036 | tRNA | 135953 | 136024 | minus | |
| 40863775 | Ocimum tenuiflorum | trnA-UGC | FPH81_pgt034 | tRNA | 131978 | 132855 | minus | |
| 40863772 | Ocimum tenuiflorum | trnR-ACG | FPH81_pgt029 | tRNA | 106327 | 106400 | plus | |
| 40863769 | Ocimum tenuiflorum | trnI-GAU | FPH81_pgt027 | tRNA | 100573 | 101589 | plus | |
| 40863768 | Ocimum tenuiflorum | trnH-GUG | FPH81_pgt001 | tRNA | 5 | 79 | minus | |
| 40863766 | Ocimum tenuiflorum | trnI-CAU | FPH81_pgt038 | tRNA | 149609 | 149682 | plus | |
| 40863763 | Ocimum tenuiflorum | trnL-CAA | FPH81_pgt037 | tRNA | 142019 | 142099 | plus | |
| 40863758 | Ocimum tenuiflorum | trnI-GAU | FPH81_pgt035 | tRNA | 132920 | 133936 | minus | |
| 40863756 | Ocimum tenuiflorum | trnR-ACG | FPH81_pgt033 | tRNA | 128109 | 128182 | minus | |
| 40863755 | Ocimum tenuiflorum | trnN-GUU | FPH81_pgt032 | tRNA | 127470 | 127542 | plus | |
| 40863744 | Ocimum tenuiflorum | trnL-UAG | FPH81_pgt031 | tRNA | 111979 | 112058 | plus | |
| 40863740 | Ocimum tenuiflorum | trnN-GUU | FPH81_pgt030 | tRNA | 106967 | 107039 | minus | |
| 40863738 | Ocimum tenuiflorum | trnA-UGC | FPH81_pgt028 | tRNA | 101654 | 102531 | plus | |
| 40863736 | Ocimum tenuiflorum | trnV-GAC | FPH81_pgt026 | tRNA | 98485 | 98556 | plus | |
| 40863733 | Ocimum tenuiflorum | trnL-CAA | FPH81_pgt025 | tRNA | 92410 | 92490 | minus | |
| 40863730 | Ocimum tenuiflorum | trnI-CAU | FPH81_pgt024 | tRNA | 84827 | 84900 | minus | |
| 40863706 | Ocimum tenuiflorum | trnP-GGG | FPH81_pgt023 | tRNA | 65737 | 65807 | minus | |
| 40863705 | Ocimum tenuiflorum | trnP-UGG | FPH81_pgt022 | tRNA | 65735 | 65808 | minus | |
| 40863704 | Ocimum tenuiflorum | trnW-CCA | FPH81_pgt021 | tRNA | 65493 | 65566 | minus | |
| 40863689 | Ocimum tenuiflorum | trnM-CAU | FPH81_pgt020 | tRNA | 51452 | 51523 | plus | |
| 40863688 | Ocimum tenuiflorum | trnV-UAC | FPH81_pgt019 | tRNA | 50620 | 51270 | minus | |
| 40863684 | Ocimum tenuiflorum | trnF-GAA | FPH81_pgt018 | tRNA | 47264 | 47336 | plus | |
| 40863683 | Ocimum tenuiflorum | trnL-UAA | FPH81_pgt017 | tRNA | 46434 | 46988 | plus | |
| 40863682 | Ocimum tenuiflorum | trnT-UGU | FPH81_pgt016 | tRNA | 45672 | 45744 | minus | |
| 40863680 | Ocimum tenuiflorum | trnS-GGA | FPH81_pgt015 | tRNA | 44299 | 44385 | plus | |
| 40863675 | Ocimum tenuiflorum | trnfM-CAU | FPH81_pgt014 | tRNA | 35585 | 35658 | minus | |
| 40863674 | Ocimum tenuiflorum | trnG-UCC | FPH81_pgt013 | tRNA | 35344 | 35414 | plus | |
| 40863672 | Ocimum tenuiflorum | trnS-UGA | FPH81_pgt012 | tRNA | 34485 | 34576 | minus | |
| 40863669 | Ocimum tenuiflorum | trnM-CAU | FPH81_pgt011 | tRNA | 30486 | 30544 | plus | |
| 40863668 | Ocimum tenuiflorum | trnT-GGU | FPH81_pgt010 | tRNA | 30480 | 30551 | plus | |
| 40863667 | Ocimum tenuiflorum | trnE-UUC | FPH81_pgt009 | tRNA | 29798 | 29870 | minus | |
| 40863666 | Ocimum tenuiflorum | trnY-GUA | FPH81_pgt008 | tRNA | 29654 | 29737 | minus | |
| 40863665 | Ocimum tenuiflorum | trnD-GUC | FPH81_pgt007 | tRNA | 29464 | 29537 | minus | |
| 40863662 | Ocimum tenuiflorum | trnC-GCA | FPH81_pgt006 | tRNA | 27504 | 27584 | plus | |
| 40863653 | Ocimum tenuiflorum | trnR-UCU | FPH81_pgt005 | tRNA | 9435 | 9506 | plus | |
| 40863652 | Ocimum tenuiflorum | trnS-GCU | FPH81_pgt004 | tRNA | 7646 | 7733 | minus | |
| 40863649 | Ocimum tenuiflorum | trnQ-UUG | FPH81_pgt003 | tRNA | 6369 | 6440 | minus |
miRNA Information
| Acc. No. | Species Name | Sequence length | Total pre-miRNA | Sequence |
|---|---|---|---|---|
| MF468201.1 | Ocimum tenuiflorum | 751 | 10 Details |
RNA-Seq Information
| Experiment Acc. | Species Name | SRA Study | SRA Run | Treatment | Development Stage | Tissue | Data Accesss |
|---|---|---|---|---|---|---|---|
| SRX10464861 | Ocimum tenuiflorum | SRP312538 | SRR14090688 | chloroplast genome | |||
| SRX10243622 | Ocimum tenuiflorum | SRP309435 | SRR13862647 | chloroplast genome | |||
| SRX2917198 | Ocimum tenuiflorum | SRP109145 | SRR5681938 | drought | leaf | ||
| SRX2917191 | Ocimum tenuiflorum | SRP109145 | SRR5681928 | Salt | leaf | ||
| SRX2917189 | Ocimum tenuiflorum | SRP109145 | SRR5681927 | Flood | leaf | ||
| SRX2917188 | Ocimum tenuiflorum | SRP109145 | SRR5681926 | Cold | leaf | ||
| SRX2916800 | Ocimum tenuiflorum | SRP109145 | SRR5681516 | Control | leaf | ||
| SRX2916759 | Ocimum tenuiflorum | SRP109144 | SRR5681434 | Heat | leaf | ||
| SRX835417 | Ocimum tenuiflorum | SRP051184 | SRR1744087 | Genome | |||
| SRX832016 | Ocimum tenuiflorum | SRP051184 | SRR1767353 | ||||
| SRX810861 | Ocimum tenuiflorum | SRP051184 | SRR1715044 | Genome | |||
| SRX761338 | Ocimum tenuiflorum | SRP049911 | SRR1654829 | ||||
| SRX760129 | Ocimum tenuiflorum | SRP049911 | SRR1653607 | ||||
| SRX760132 | Ocimum tenuiflorum | SRP049911 | SRR1653610 | Genome | |||
| SRX476890 | Ocimum tenuiflorum | SRP039008 | SRR1177846 |
Other non-coding Sequence Information
* If data is not displayed please access the respective tab