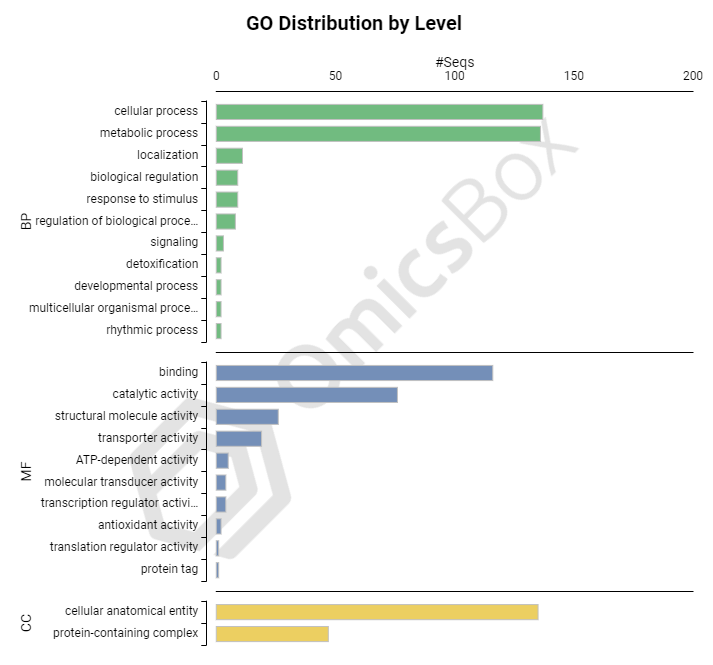
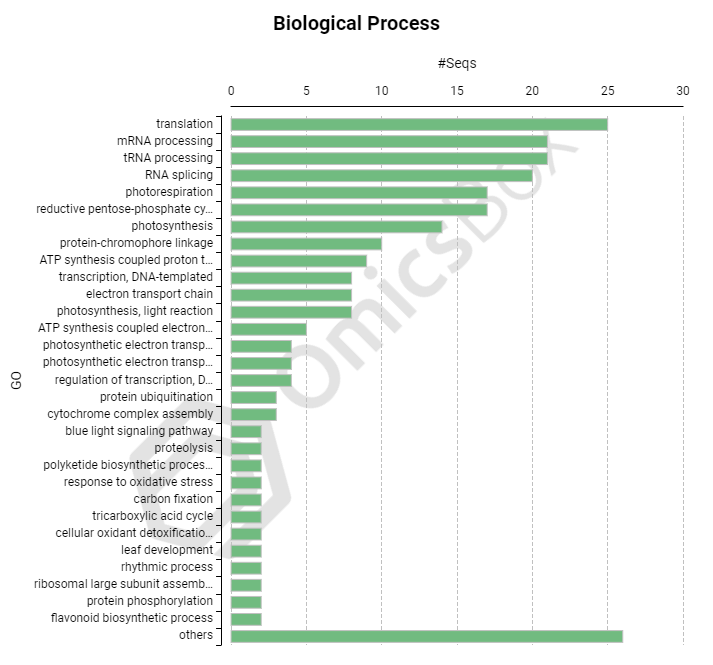
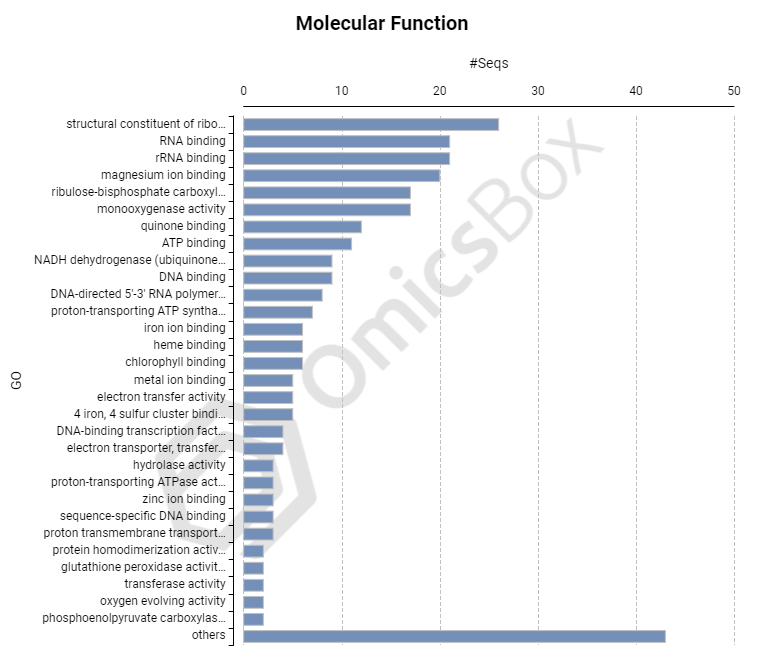
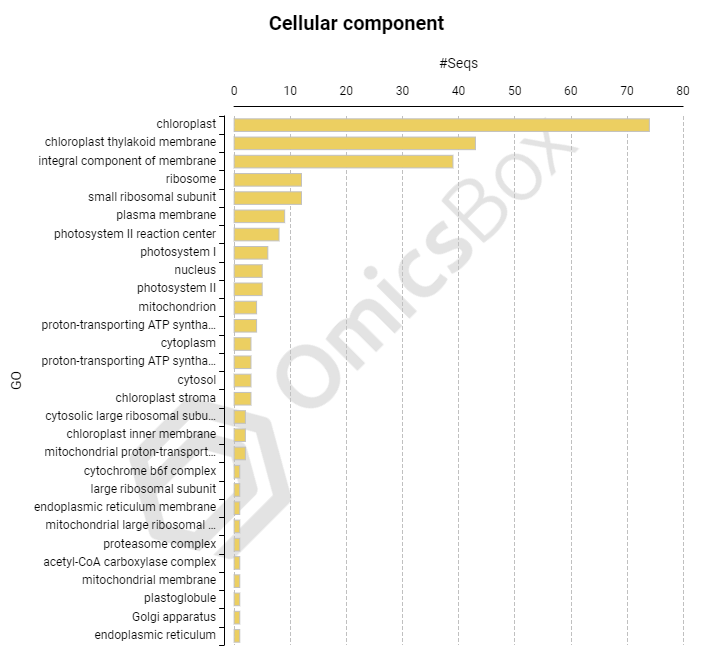
Coding Information
| Acc No. | Species Name | Description | Length | GO IDs | GO Names | Enzyme Names | InterPro GO Names | Peptide Information | BLAST | Nucleotide Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| JG648073.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 804 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG647189.1 | Tinospora cordifolia | glyceraldehyde-3-phosphate dehydrogenase, cytosolic | 748 | P:GO:0006006; P:GO:0006096; F:GO:0004365; F:GO:0050661; F:GO:0051287; C:GO:0005829; C:GO:0009536 | P:glucose metabolic process; P:glycolytic process; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity; F:NADP binding; F:NAD binding; C:cytosol; C:plastid | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating); Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG648119.1 | Tinospora cordifolia | Glutathione S-transferase | 426 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | F:glutathione transferase activity | view details | ||
| JG648523.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 730 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646331.1 | Tinospora cordifolia | thaumatin-like protein | 755 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG645770.1 | Tinospora cordifolia | ribosomal RNA small subunit methyltransferase-like | 704 | P:GO:0001708; P:GO:0006338; P:GO:0031167; P:GO:0051301; F:GO:0000179; F:GO:0003723; F:GO:0005524; F:GO:0140658; C:GO:0005730; C:GO:0005759 | P:cell fate specification; P:chromatin remodeling; P:rRNA methylation; P:cell division; F:rRNA (adenine-N6,N6-)-dimethyltransferase activity; F:RNA binding; F:ATP binding; F:ATP-dependent chromatin remodeler activity; C:nucleolus; C:mitochondrial matrix | Transferring one-carbon groups | P:rRNA processing; F:rRNA (adenine-N6,N6-)-dimethyltransferase activity | view details | ||
| JG646634.1 | Tinospora cordifolia | mediator of RNA polymerase II transcription subunit 25 | 878 | P:GO:0045944; C:GO:0005667; C:GO:0016592 | P:positive regulation of transcription by RNA polymerase II; C:transcription regulator complex; C:mediator complex | no GO terms | view details | |||
| JG647350.1 | Tinospora cordifolia | acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like | 733 | P:GO:0006633; P:GO:2001295; F:GO:0003989; F:GO:0005524; F:GO:0016740; C:GO:0009317 | P:fatty acid biosynthetic process; P:malonyl-CoA biosynthetic process; F:acetyl-CoA carboxylase activity; F:ATP binding; F:transferase activity; C:acetyl-CoA carboxylase complex | Transferases; Acetyl-CoA carboxylase | P:fatty acid biosynthetic process; F:acetyl-CoA carboxylase activity; C:acetyl-CoA carboxylase complex | view details | ||
| JG646898.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 669 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648321.1 | Tinospora cordifolia | actin-like | 873 | F:GO:0005524; C:GO:0005737; C:GO:0005856 | F:ATP binding; C:cytoplasm; C:cytoskeleton | no GO terms | view details | |||
| JG648796.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 756 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG647178.1 | Tinospora cordifolia | isocitrate lyase | 856 | P:GO:0006097; P:GO:0006099; F:GO:0004451; F:GO:0046872; C:GO:0009514 | P:glyoxylate cycle; P:tricarboxylic acid cycle; F:isocitrate lyase activity; F:metal ion binding; C:glyoxysome | Isocitrate lyase | P:carboxylic acid metabolic process; F:catalytic activity; F:isocitrate lyase activity | view details | ||
| JG648992.1 | Tinospora cordifolia | Long chain base biosynthesis protein 2a | 510 | P:GO:0046512; P:GO:0046513; F:GO:0004758; F:GO:0008483; F:GO:0030170; C:GO:0016021; C:GO:0017059 | P:sphingosine biosynthetic process; P:ceramide biosynthetic process; F:serine C-palmitoyltransferase activity; F:transaminase activity; F:pyridoxal phosphate binding; C:integral component of membrane; C:serine C-palmitoyltransferase complex | Transferring nitrogenous groups; Serine C-palmitoyltransferase | P:biosynthetic process; F:pyridoxal phosphate binding | view details | ||
| JG646017.1 | Tinospora cordifolia | cytochrome P450 81E8-like | 796 | P:GO:0009058; F:GO:0005488; F:GO:0016491 | P:biosynthetic process; F:binding; F:oxidoreductase activity | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649702.1 | Tinospora cordifolia | 60S ribosomal protein L10 | 712 | P:GO:0000027; P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646023.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 780 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648019.1 | Tinospora cordifolia | Aconitate hydratase, cytoplasmic | 746 | P:GO:0006099; P:GO:0006101; F:GO:0003994; F:GO:0046872; F:GO:0047780; F:GO:0051539; C:GO:0005739; C:GO:0005829 | P:tricarboxylic acid cycle; P:citrate metabolic process; F:aconitate hydratase activity; F:metal ion binding; F:citrate dehydratase activity; F:4 iron, 4 sulfur cluster binding; C:mitochondrion; C:cytosol | Aconitate hydratase | no GO terms | view details | ||
| JG649054.1 | Tinospora cordifolia | S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase, chloroplastic-like | 772 | P:GO:0019354; P:GO:0032259; F:GO:0004851 | P:siroheme biosynthetic process; P:methylation; F:uroporphyrin-III C-methyltransferase activity | Uroporphyrinogen-III C-methyltransferase | P:siroheme biosynthetic process; F:methyltransferase activity | view details | ||
| JG646862.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 529 | F:GO:0050660 | F:flavin adenine dinucleotide binding | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | |||
| JG645777.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 744 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646050.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 760 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG646457.1 | Tinospora cordifolia | FAD linked oxidase | 826 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650011.1 | Tinospora cordifolia | FAD linked oxidase | 741 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650547.1 | Tinospora cordifolia | probable flavin-containing monooxygenase 1 | 114 | F:GO:0000166; F:GO:0004497 | F:nucleotide binding; F:monooxygenase activity | Oxidoreductases | no IPS match | view details | ||
| JG649074.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 270 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647864.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 755 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646612.1 | Tinospora cordifolia | lysine-specific demethylase JMJ25-like | 590 | P:GO:0006357; P:GO:0033169; F:GO:0003712; F:GO:0031490; F:GO:0032454; C:GO:0000118; C:GO:0000785 | P:regulation of transcription by RNA polymerase II; P:histone H3-K9 demethylation; F:transcription coregulator activity; F:chromatin DNA binding; F:histone H3-methyl-lysine-9 demethylase activity; C:histone deacetylase complex; C:chromatin | P:histone H3-K9 demethylation; F:histone H3-methyl-lysine-9 demethylase activity | view details | |||
| MG946871.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1410 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG650915.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 701 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650502.1 | Tinospora cordifolia | Corytuberine synthase | 842 | P:GO:0009058; P:GO:0071704; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:biosynthetic process; P:organic substance metabolic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650636.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 801 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650705.1 | Tinospora cordifolia | laccase-7-like | 890 | P:GO:0046274; F:GO:0005507; F:GO:0052716; C:GO:0048046 | P:lignin catabolic process; F:copper ion binding; F:hydroquinone:oxygen oxidoreductase activity; C:apoplast | Laccase | F:copper ion binding | view details | ||
| JG650427.1 | Tinospora cordifolia | Coiled-coil domain-containing protein | 751 | P:GO:0032469; F:GO:0005509; C:GO:0005783; C:GO:0016021 | P:endoplasmic reticulum calcium ion homeostasis; F:calcium ion binding; C:endoplasmic reticulum; C:integral component of membrane | P:endoplasmic reticulum calcium ion homeostasis; F:calcium ion binding; C:endoplasmic reticulum | view details | |||
| JG645702.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 806 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649674.1 | Tinospora cordifolia | ---NA--- | 54 | no IPS match | view details | |||||
| JG645787.1 | Tinospora cordifolia | rRNA-processing protein UTP23 homolog isoform X1 | 725 | P:GO:0042254; C:GO:0016020 | P:ribosome biogenesis; C:membrane | C:small-subunit processome | view details | |||
| JG649004.1 | Tinospora cordifolia | probable mannitol dehydrogenase | 270 | P:GO:0009809; F:GO:0008270; F:GO:0045551 | P:lignin biosynthetic process; F:zinc ion binding; F:cinnamyl-alcohol dehydrogenase activity | Cinnamyl-alcohol dehydrogenase | no GO terms | view details | ||
| JG650282.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 813 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649156.1 | Tinospora cordifolia | dynamin-2A-like | 762 | F:GO:0003924; F:GO:0005525; F:GO:0008017; C:GO:0005737; C:GO:0005874; C:GO:0016020 | F:GTPase activity; F:GTP binding; F:microtubule binding; C:cytoplasm; C:microtubule; C:membrane | Acting on acid anhydrides | no GO terms | view details | ||
| JG648045.1 | Tinospora cordifolia | PKS ER domain-containing protein | 721 | F:GO:0008270; F:GO:0016491 | F:zinc ion binding; F:oxidoreductase activity | Oxidoreductases | no GO terms | view details | ||
| JG646930.1 | Tinospora cordifolia | endochitinase CH25-like | 158 | P:GO:0000272; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:polysaccharide catabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | no IPS match | view details | ||
| JG646939.1 | Tinospora cordifolia | eukaryotic translation initiation factor 3 subunit A-like | 571 | P:GO:0001732; P:GO:0002188; F:GO:0003729; F:GO:0003743; C:GO:0000502; C:GO:0016021; C:GO:0016282; C:GO:0033290; C:GO:0043614; C:GO:0071540; C:GO:0071541 | P:formation of cytoplasmic translation initiation complex; P:translation reinitiation; F:mRNA binding; F:translation initiation factor activity; C:proteasome complex; C:integral component of membrane; C:eukaryotic 43S preinitiation complex; C:eukaryotic 48S preinitiation complex; C:multi-eIF complex; C:eukaryotic translation initiation factor 3 complex, eIF3e; C:eukaryotic translation initiation factor 3 complex, eIF3m | C:eukaryotic translation initiation factor 3 complex | view details | |||
| MH569072.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 375 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG646920.1 | Tinospora cordifolia | UDP-glycosyltransferase 87A1-like | 606 | F:GO:0016740 | F:transferase activity | Transferases | no GO terms | view details | ||
| JG646491.1 | Tinospora cordifolia | probable LRR receptor-like serine/threonine-protein kinase At1g14390 | 730 | P:GO:0046777; F:GO:0004672; C:GO:0005886 | P:protein autophosphorylation; F:protein kinase activity; C:plasma membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:transmembrane receptor protein tyrosine kinase activity; F:ATP binding | view details | ||
| JG650619.1 | Tinospora cordifolia | serine hydroxymethyltransferase 4 | 732 | P:GO:0006565; P:GO:0019264; P:GO:0032259; P:GO:0035999; P:GO:0046655; F:GO:0004372; F:GO:0008168; F:GO:0008270; F:GO:0030170; F:GO:0070905; C:GO:0005737 | P:L-serine catabolic process; P:glycine biosynthetic process from serine; P:methylation; P:tetrahydrofolate interconversion; P:folic acid metabolic process; F:glycine hydroxymethyltransferase activity; F:methyltransferase activity; F:zinc ion binding; F:pyridoxal phosphate binding; F:serine binding; C:cytoplasm | Glycine hydroxymethyltransferase; Transferring one-carbon groups | P:glycine biosynthetic process from serine; P:tetrahydrofolate interconversion; F:catalytic activity; F:glycine hydroxymethyltransferase activity; F:pyridoxal phosphate binding | view details | ||
| JG647027.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 783 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650859.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 773 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647325.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP) | 820 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG650311.1 | Tinospora cordifolia | protein disulfide-isomerase | 803 | P:GO:0006457; P:GO:0034976; F:GO:0003756; C:GO:0005788 | P:protein folding; P:response to endoplasmic reticulum stress; F:protein disulfide isomerase activity; C:endoplasmic reticulum lumen | Protein disulfide-isomerase | F:protein disulfide isomerase activity | view details | ||
| JG649286.1 | Tinospora cordifolia | dynamin-2A-like isoform X2 | 851 | F:GO:0003924; F:GO:0005525; F:GO:0008017; C:GO:0005737; C:GO:0005874; C:GO:0016020 | F:GTPase activity; F:GTP binding; F:microtubule binding; C:cytoplasm; C:microtubule; C:membrane | Acting on acid anhydrides | no GO terms | view details | ||
| JG650475.1 | Tinospora cordifolia | 4-coumarate--CoA ligase-like 5 | 872 | P:GO:0009611; P:GO:0009617; P:GO:0009695; P:GO:0009698; P:GO:0009753; F:GO:0016207; C:GO:0005777; C:GO:0016021 | P:response to wounding; P:response to bacterium; P:jasmonic acid biosynthetic process; P:phenylpropanoid metabolic process; P:response to jasmonic acid; F:4-coumarate-CoA ligase activity; C:peroxisome; C:integral component of membrane | 4-coumarate--CoA ligase | no GO terms | view details | ||
| JG645520.1 | Tinospora cordifolia | aspartic proteinase A1 | 648 | P:GO:0006508; P:GO:0006629; F:GO:0004190 | P:proteolysis; P:lipid metabolic process; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG647795.1 | Tinospora cordifolia | cell division cycle protein 48 homolog | 743 | P:GO:0030433; P:GO:0030970; P:GO:0051228; P:GO:0051301; P:GO:0071712; P:GO:0097352; F:GO:0005524; F:GO:0008237; F:GO:0016887; F:GO:0031593; C:GO:0005634; C:GO:0005829; C:GO:0016021; C:GO:0034098 | P:ubiquitin-dependent ERAD pathway; P:retrograde protein transport, ER to cytosol; P:mitotic spindle disassembly; P:cell division; P:ER-associated misfolded protein catabolic process; P:autophagosome maturation; F:ATP binding; F:metallopeptidase activity; F:ATP hydrolysis activity; F:polyubiquitin modification-dependent protein binding; C:nucleus; C:cytosol; C:integral component of membrane; C:VCP-NPL4-UFD1 AAA ATPase complex | Acting on acid anhydrides; Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| JG649347.1 | Tinospora cordifolia | endoplasmin homolog | 797 | P:GO:0006457; P:GO:0009306; P:GO:0009414; P:GO:0009651; P:GO:0009934; P:GO:0010075; P:GO:0034976; F:GO:0005524; F:GO:0016887; F:GO:0051082; C:GO:0005783; C:GO:0048471 | P:protein folding; P:protein secretion; P:response to water deprivation; P:response to salt stress; P:regulation of meristem structural organization; P:regulation of meristem growth; P:response to endoplasmic reticulum stress; F:ATP binding; F:ATP hydrolysis activity; F:unfolded protein binding; C:endoplasmic reticulum; C:perinuclear region of cytoplasm | Acting on acid anhydrides | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; F:unfolded protein binding | view details | ||
| JG649875.1 | Tinospora cordifolia | basic endochitinase-like | 389 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646174.1 | Tinospora cordifolia | Polyphenol oxidase protein | 513 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | no GO terms | view details | ||
| JG648827.1 | Tinospora cordifolia | S-adenosylmethionine synthetase | 437 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG649464.1 | Tinospora cordifolia | transaldolase isoform X2 | 771 | P:GO:0005975; P:GO:0006098; F:GO:0004801; C:GO:0005737 | P:carbohydrate metabolic process; P:pentose-phosphate shunt; F:transaldolase activity; C:cytoplasm | Transaldolase | P:carbohydrate metabolic process; P:pentose-phosphate shunt; F:transaldolase activity; C:cytoplasm | view details | ||
| JG647390.1 | Tinospora cordifolia | Purine permease | 844 | P:GO:0015860; P:GO:1904823; F:GO:0005345; F:GO:0015211; C:GO:0016021 | P:purine nucleoside transmembrane transport; P:purine nucleobase transmembrane transport; F:purine nucleobase transmembrane transporter activity; F:purine nucleoside transmembrane transporter activity; C:integral component of membrane | Translocases | F:purine nucleoside transmembrane transporter activity; C:integral component of membrane | view details | ||
| JG645818.1 | Tinospora cordifolia | triosephosphate isomerase, cytosolic | 809 | P:GO:0006094; P:GO:0006096; P:GO:0019563; P:GO:0046166; F:GO:0004807; C:GO:0005829 | P:gluconeogenesis; P:glycolytic process; P:glycerol catabolic process; P:glyceraldehyde-3-phosphate biosynthetic process; F:triose-phosphate isomerase activity; C:cytosol | Triose-phosphate isomerase | P:glycolytic process; F:triose-phosphate isomerase activity | view details | ||
| JG647450.1 | Tinospora cordifolia | 3-dehydroquinate synthase | 718 | P:GO:0008652; P:GO:0009073; P:GO:0009423; F:GO:0003856; F:GO:0042802; F:GO:0046872; F:GO:0051287; C:GO:0009507 | P:cellular amino acid biosynthetic process; P:aromatic amino acid family biosynthetic process; P:chorismate biosynthetic process; F:3-dehydroquinate synthase activity; F:identical protein binding; F:metal ion binding; F:NAD binding; C:chloroplast | 3-dehydroquinate synthase | no GO terms | view details | ||
| JG648661.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 849 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| MS687776.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1569 | P:GO:0006259; P:GO:0006974; P:GO:0009058; P:GO:0051276; P:GO:0098542; F:GO:0000166; F:GO:0004386; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:DNA metabolic process; P:cellular response to DNA damage stimulus; P:biosynthetic process; P:chromosome organization; P:defense response to other organism; F:nucleotide binding; F:helicase activity; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647238.1 | Tinospora cordifolia | Nucleolar protein 56 | 820 | P:GO:0042254; F:GO:0030515; C:GO:0005730; C:GO:0031428; C:GO:0032040 | P:ribosome biogenesis; F:snoRNA binding; C:nucleolus; C:box C/D RNP complex; C:small-subunit processome | F:snoRNA binding; C:box C/D RNP complex; C:small-subunit processome | view details | |||
| JG647451.1 | Tinospora cordifolia | CBS domain-containing protein CBSCBSPB3-like | 609 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG647103.1 | Tinospora cordifolia | L-ascorbate peroxidase, cytosolic | 753 | P:GO:0000302; P:GO:0034599; P:GO:0042744; P:GO:0098869; F:GO:0016688; F:GO:0020037; F:GO:0046872; C:GO:0009507 | P:response to reactive oxygen species; P:cellular response to oxidative stress; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:L-ascorbate peroxidase activity; F:heme binding; F:metal ion binding; C:chloroplast | L-ascorbate peroxidase | P:response to oxidative stress; P:cellular response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG647044.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 874 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650995.1 | Tinospora cordifolia | NADP-dependent malic enzyme isoform X2 | 808 | P:GO:0006108; F:GO:0004471; F:GO:0046872; F:GO:0051287 | P:malate metabolic process; F:malate dehydrogenase (decarboxylating) (NAD+) activity; F:metal ion binding; F:NAD binding | Malate dehydrogenase (oxaloacetate-decarboxylating); Malate dehydrogenase (decarboxylating) | F:malic enzyme activity; F:malate dehydrogenase (decarboxylating) (NAD+) activity; F:NAD binding | view details | ||
| JG649715.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 273 | P:GO:0019438; P:GO:1901362; F:GO:0005506; F:GO:0020037; F:GO:0047087; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:iron ion binding; F:heme binding; F:protopine 6-monooxygenase activity; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Protopine 6-monooxygenase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649966.1 | Tinospora cordifolia | probable fructokinase-4 | 493 | P:GO:0006000; P:GO:0019252; P:GO:0046835; F:GO:0008865; C:GO:0005829 | P:fructose metabolic process; P:starch biosynthetic process; P:carbohydrate phosphorylation; F:fructokinase activity; C:cytosol | Fructokinase; Hexokinase | no GO terms | view details | ||
| JG648645.1 | Tinospora cordifolia | 2-oxoglutarate-dependent dioxygenase DAO-like | 803 | P:GO:0009852; P:GO:0010252; P:GO:1901576; F:GO:0016707; F:GO:0046872; F:GO:0050302 | P:auxin catabolic process; P:auxin homeostasis; P:organic substance biosynthetic process; F:gibberellin 3-beta-dioxygenase activity; F:metal ion binding; F:indole-3-acetaldehyde oxidase activity | Gibberellin 3-beta-dioxygenase; Aryl-aldehyde oxidase; Indole-3-acetaldehyde oxidase; Aldehyde oxidase | no GO terms | view details | ||
| JG647684.1 | Tinospora cordifolia | S-adenosylmethionine synthase 1 | 849 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG645703.1 | Tinospora cordifolia | putative E3 ubiquitin-protein ligase XBAT31 | 812 | P:GO:0009408; P:GO:0036211; F:GO:0016740; F:GO:0140096 | P:response to heat; P:protein modification process; F:transferase activity; F:catalytic activity, acting on a protein | Transferases | F:protein binding | view details | ||
| JG647498.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 823 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649370.1 | Tinospora cordifolia | NADPH--cytochrome P450 reductase | 752 | F:GO:0003958; F:GO:0010181; F:GO:0050660; F:GO:0050661; C:GO:0005789; C:GO:0005829; C:GO:0016021 | F:NADPH-hemoprotein reductase activity; F:FMN binding; F:flavin adenine dinucleotide binding; F:NADP binding; C:endoplasmic reticulum membrane; C:cytosol; C:integral component of membrane | NADPH--hemoprotein reductase | F:oxidoreductase activity | view details | ||
| JG649309.1 | Tinospora cordifolia | Cytochrome p450 | 797 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645743.1 | Tinospora cordifolia | polyubiquitin 3 | 693 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | F:protein binding | view details | |||
| MG564657.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 505 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG647024.1 | Tinospora cordifolia | 3-phosphoshikimate 1-carboxyvinyltransferase 2 | 718 | P:GO:0008652; P:GO:0009073; P:GO:0009423; F:GO:0003866 | P:cellular amino acid biosynthetic process; P:aromatic amino acid family biosynthetic process; P:chorismate biosynthetic process; F:3-phosphoshikimate 1-carboxyvinyltransferase activity | 3-phosphoshikimate 1-carboxyvinyltransferase | F:catalytic activity; F:transferase activity, transferring alkyl or aryl (other than methyl) groups | view details | ||
| JG648632.1 | Tinospora cordifolia | endoglucanase 12 | 811 | P:GO:0030245; F:GO:0008810; C:GO:0016021 | P:cellulose catabolic process; F:cellulase activity; C:integral component of membrane | Cellulase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649405.1 | Tinospora cordifolia | ornithine aminotransferase, mitochondrial | 702 | P:GO:0006593; P:GO:0010121; P:GO:0019544; P:GO:0042538; P:GO:0042742; P:GO:0051646; P:GO:0055129; F:GO:0004587; F:GO:0030170; F:GO:0042802; F:GO:0050155; C:GO:0005739 | P:ornithine catabolic process; P:arginine catabolic process to proline via ornithine; P:arginine catabolic process to glutamate; P:hyperosmotic salinity response; P:defense response to bacterium; P:mitochondrion localization; P:L-proline biosynthetic process; F:ornithine-oxo-acid transaminase activity; F:pyridoxal phosphate binding; F:identical protein binding; F:ornithine(lysine) transaminase activity; C:mitochondrion | Ornithine aminotransferase | F:catalytic activity; F:ornithine-oxo-acid transaminase activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG649024.1 | Tinospora cordifolia | thaumatin-like protein | 292 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647743.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A-like | 76 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | no IPS match | view details | |||
| JG648449.1 | Tinospora cordifolia | BURP domain protein RD22-like | 838 | no GO terms | view details | |||||
| JG650792.1 | Tinospora cordifolia | monodehydroascorbate reductase | 822 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG647510.1 | Tinospora cordifolia | 60S ribosomal protein L5 | 754 | P:GO:0000027; P:GO:0006412; F:GO:0003735; F:GO:0008097; C:GO:0005634; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:structural constituent of ribosome; F:5S rRNA binding; C:nucleus; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; F:5S rRNA binding; C:ribosome | view details | |||
| JG645857.1 | Tinospora cordifolia | Pyrophosphate-energized vacuolar membrane proton pump | 824 | P:GO:1902600; F:GO:0004427; F:GO:0009678; C:GO:0016021 | P:proton transmembrane transport; F:inorganic diphosphatase activity; F:pyrophosphate hydrolysis-driven proton transmembrane transporter activity; C:integral component of membrane | H(+)-exporting diphosphatase; Inorganic diphosphatase | P:proton transmembrane transport; F:inorganic diphosphatase activity; F:pyrophosphate hydrolysis-driven proton transmembrane transporter activity; C:membrane | view details | ||
| JG646678.1 | Tinospora cordifolia | cytochrome P450 | 822 | P:GO:0009058; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649872.1 | Tinospora cordifolia | Acetate/butyrate--CoA ligase AAE7, peroxisomal | 626 | P:GO:0006083; P:GO:0006097; P:GO:0009698; P:GO:0019605; F:GO:0003987; F:GO:0016207; F:GO:0047760; F:GO:0106290; C:GO:0005777 | P:acetate metabolic process; P:glyoxylate cycle; P:phenylpropanoid metabolic process; P:butyrate metabolic process; F:acetate-CoA ligase activity; F:4-coumarate-CoA ligase activity; F:butyrate-CoA ligase activity; F:trans-cinnamate-CoA ligase activity; C:peroxisome | 4-coumarate--CoA ligase; Medium-chain acyl-CoA ligase | no GO terms | view details | ||
| JG650580.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 821 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649829.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 807 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG647865.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 438 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650763.1 | Tinospora cordifolia | protein DETOXIFICATION 29 | 866 | P:GO:1990961; F:GO:0015297; F:GO:0042910; C:GO:0005774; C:GO:0016021 | P:xenobiotic detoxification by transmembrane export across the plasma membrane; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:vacuolar membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:membrane | view details | ||
| JG649291.1 | Tinospora cordifolia | ---NA--- | 59 | no IPS match | view details | |||||
| JG650669.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 812 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| MK244348.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1419 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG646882.1 | Tinospora cordifolia | 6-phosphogluconate dehydrogenase | 551 | P:GO:0006098; P:GO:0019521; F:GO:0004616; F:GO:0050661 | P:pentose-phosphate shunt; P:D-gluconate metabolic process; F:phosphogluconate dehydrogenase (decarboxylating) activity; F:NADP binding | Phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | F:phosphogluconate dehydrogenase (decarboxylating) activity; F:NADP binding | view details | ||
| JG645723.1 | Tinospora cordifolia | Corytuberine synthase | 759 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648787.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 542 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645809.1 | Tinospora cordifolia | auxin response factor 2A-like | 719 | P:GO:0006355; P:GO:0009725; P:GO:0009734; F:GO:0003677; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:response to hormone; P:auxin-activated signaling pathway; F:DNA binding; C:nucleus | P:regulation of transcription, DNA-templated; P:response to hormone; F:DNA binding | view details | |||
| JG650279.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 822 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650863.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 812 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649058.1 | Tinospora cordifolia | serine/threonine-protein phosphatase PP1 isoform X1 | 780 | P:GO:0006470; F:GO:0003729; F:GO:0017018; C:GO:0005634; C:GO:0005737; C:GO:0015934 | P:protein dephosphorylation; F:mRNA binding; F:myosin phosphatase activity; C:nucleus; C:cytoplasm; C:large ribosomal subunit | Protein-serine/threonine phosphatase | F:hydrolase activity | view details | ||
| JG649761.1 | Tinospora cordifolia | ribosome biogenesis protein NSA2 homolog | 541 | P:GO:0000460; P:GO:0000470; C:GO:0005730; C:GO:0030687 | P:maturation of 5.8S rRNA; P:maturation of LSU-rRNA; C:nucleolus; C:preribosome, large subunit precursor | no GO terms | view details | |||
| JG646514.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 135 | P:GO:0006952; P:GO:0009607; P:GO:0009738; P:GO:0043086; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:defense response; P:response to biotic stimulus; P:abscisic acid-activated signaling pathway; P:negative regulation of catalytic activity; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | no IPS match | view details | |||
| JG650119.1 | Tinospora cordifolia | basic endochitinase | 785 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647408.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase 1 | 806 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG648972.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 605 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Acting on acid anhydrides; Lyases | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG646702.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 745 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG650141.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 737 | P:GO:0005975; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647545.1 | Tinospora cordifolia | aspartic proteinase-like | 706 | P:GO:0006508; P:GO:0006629; F:GO:0004190 | P:proteolysis; P:lipid metabolic process; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG648845.1 | Tinospora cordifolia | glutathione S-transferase L3-like | 342 | P:GO:0006749; P:GO:0010731; F:GO:0004364 | P:glutathione metabolic process; P:protein glutathionylation; F:glutathione transferase activity | Glutathione transferase | F:glutathione transferase activity | view details | ||
| JG647373.1 | Tinospora cordifolia | U-box domain-containing protein 16 | 754 | P:GO:0007166; P:GO:0016567; F:GO:0004842; F:GO:0016746; F:GO:0016874; C:GO:0005634; C:GO:0005737 | P:cell surface receptor signaling pathway; P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:acyltransferase activity; F:ligase activity; C:nucleus; C:cytoplasm | Acyltransferases; Ligases | P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:protein binding | view details | ||
| JG647568.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 801 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650982.1 | Tinospora cordifolia | oxalate--CoA ligase | 785 | P:GO:0006631; P:GO:0009698; P:GO:0010030; P:GO:0010214; P:GO:0033611; P:GO:0050832; F:GO:0016207; F:GO:0031956; F:GO:0050203; C:GO:0005737 | P:fatty acid metabolic process; P:phenylpropanoid metabolic process; P:positive regulation of seed germination; P:seed coat development; P:oxalate catabolic process; P:defense response to fungus; F:4-coumarate-CoA ligase activity; F:medium-chain fatty acid-CoA ligase activity; F:oxalate-CoA ligase activity; C:cytoplasm | 4-coumarate--CoA ligase; Oxalate--CoA ligase; Medium-chain acyl-CoA ligase | no GO terms | view details | ||
| JG647942.1 | Tinospora cordifolia | protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X2 | 641 | P:GO:0006468; P:GO:0006470; F:GO:0004672; F:GO:0005524; F:GO:0017018; F:GO:0046872; C:GO:0016021 | P:protein phosphorylation; P:protein dephosphorylation; F:protein kinase activity; F:ATP binding; F:myosin phosphatase activity; F:metal ion binding; C:integral component of membrane | Transferring phosphorus-containing groups; Protein-serine/threonine phosphatase | no GO terms | view details | ||
| JG647287.1 | Tinospora cordifolia | glycine-rich domain-containing protein 1 | 405 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG647634.1 | Tinospora cordifolia | subtilisin-like protease SBT1.4 | 860 | P:GO:0006508; P:GO:0009610; F:GO:0004252 | P:proteolysis; P:response to symbiotic fungus; F:serine-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type endopeptidase activity; F:serine-type peptidase activity | view details | ||
| JG650256.1 | Tinospora cordifolia | subtilisin-like protease SBT1.4 | 766 | P:GO:0006508; P:GO:0009610; F:GO:0004252 | P:proteolysis; P:response to symbiotic fungus; F:serine-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type endopeptidase activity; F:serine-type peptidase activity | view details | ||
| JG649025.1 | Tinospora cordifolia | ---NA--- | 204 | no IPS match | view details | |||||
| JG649799.1 | Tinospora cordifolia | g-type lectin s-receptor-like serine/threonine-protein kinase | 790 | F:GO:0004672; C:GO:0016020 | F:protein kinase activity; C:membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG648080.1 | Tinospora cordifolia | chaperonin CPN60-2, mitochondrial | 777 | P:GO:0042026; F:GO:0005524; F:GO:0016887; C:GO:0005739 | P:protein refolding; F:ATP binding; F:ATP hydrolysis activity; C:mitochondrion | Acting on acid anhydrides | P:protein refolding; F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG650964.1 | Tinospora cordifolia | FAD linked oxidase | 779 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648677.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 840 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | P:proteolysis; F:aspartic-type endopeptidase activity | view details | |||
| JG649460.1 | Tinospora cordifolia | isocitrate dehydrogenase [NADP] | 832 | P:GO:0006099; P:GO:0006102; P:GO:0006739; F:GO:0000287; F:GO:0004450; F:GO:0051287; C:GO:0005739 | P:tricarboxylic acid cycle; P:isocitrate metabolic process; P:NADP metabolic process; F:magnesium ion binding; F:isocitrate dehydrogenase (NADP+) activity; F:NAD binding; C:mitochondrion | Isocitrate dehydrogenase (NADP(+)) | P:isocitrate metabolic process; F:isocitrate dehydrogenase (NADP+) activity | view details | ||
| JG650605.1 | Tinospora cordifolia | linamarin synthase 2-like | 852 | P:GO:0009718; F:GO:0047213; F:GO:0080043; F:GO:0080044 | P:anthocyanin-containing compound biosynthetic process; F:anthocyanidin 3-O-glucosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Anthocyanidin 3-O-glucosyltransferase | no GO terms | view details | ||
| JG648553.1 | Tinospora cordifolia | 60S ribosomal protein L10 | 670 | P:GO:0000027; P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647172.1 | Tinospora cordifolia | class I chitinase | 304 | P:GO:0000272; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061; C:GO:0016021 | P:polysaccharide catabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding; C:integral component of membrane | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646584.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 754 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464; F:GO:0071949 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity; F:FAD binding | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG650197.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2 | 618 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG649035.1 | Tinospora cordifolia | monodehydroascorbate reductase | 472 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG647029.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 800 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG645551.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 661 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649722.1 | Tinospora cordifolia | 2,3-dimethylmalate lyase isoform X1 | 678 | F:GO:0004451 | F:isocitrate lyase activity | Isocitrate lyase | F:catalytic activity | view details | ||
| JG650085.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 783 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648210.1 | Tinospora cordifolia | AAA-ATPase At3g50940-like | 308 | F:GO:0005524; F:GO:0016887 | F:ATP binding; F:ATP hydrolysis activity | Acting on acid anhydrides | no GO terms | view details | ||
| JG648862.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 870 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647270.1 | Tinospora cordifolia | 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 | 809 | P:GO:0006574; F:GO:0003860 | P:valine catabolic process; F:3-hydroxyisobutyryl-CoA hydrolase activity | 3-hydroxyisobutyryl-CoA hydrolase | F:3-hydroxyisobutyryl-CoA hydrolase activity | view details | ||
| JG650799.1 | Tinospora cordifolia | phosphatidylinositol 4-phosphate 5-kinase 9-like | 771 | P:GO:0005975; P:GO:0006520; P:GO:0016310; P:GO:0046854; F:GO:0005524; F:GO:0016308; C:GO:0005634; C:GO:0005829; C:GO:0005886 | P:carbohydrate metabolic process; P:cellular amino acid metabolic process; P:phosphorylation; P:phosphatidylinositol phosphate biosynthetic process; F:ATP binding; F:1-phosphatidylinositol-4-phosphate 5-kinase activity; C:nucleus; C:cytosol; C:plasma membrane | 1-phosphatidylinositol-4-phosphate 5-kinase | no GO terms | view details | ||
| JG647747.1 | Tinospora cordifolia | transaldolase isoform X1 | 677 | P:GO:0005975; P:GO:0009052; F:GO:0004801; C:GO:0005737 | P:carbohydrate metabolic process; P:pentose-phosphate shunt, non-oxidative branch; F:transaldolase activity; C:cytoplasm | Transaldolase | P:carbohydrate metabolic process | view details | ||
| JG650542.1 | Tinospora cordifolia | pectinesterase 2 | 821 | P:GO:0042545; P:GO:0043086; P:GO:0045490; F:GO:0030599; F:GO:0045330; F:GO:0046910; C:GO:0005576 | P:cell wall modification; P:negative regulation of catalytic activity; P:pectin catabolic process; F:pectinesterase activity; F:aspartyl esterase activity; F:pectinesterase inhibitor activity; C:extracellular region | Pectinesterase | P:cell wall modification; F:enzyme inhibitor activity; F:pectinesterase activity | view details | ||
| LY340342.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 990 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG647137.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 815 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG646306.1 | Tinospora cordifolia | LRR receptor-like serine threonine- kinase GSO1 | 762 | C:GO:0016020 | C:membrane | F:protein binding | view details | |||
| JG650523.1 | Tinospora cordifolia | basic endochitinase | 793 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646543.1 | Tinospora cordifolia | basic endochitinase-like | 609 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646597.1 | Tinospora cordifolia | 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 | 808 | P:GO:0008299; P:GO:0016126; F:GO:0004420; C:GO:0005778; C:GO:0005789 | P:isoprenoid biosynthetic process; P:sterol biosynthetic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; C:peroxisomal membrane; C:endoplasmic reticulum membrane | Hydroxymethylglutaryl-CoA reductase (NADPH) | P:coenzyme A metabolic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| DQ353860.2 | Tinospora cordifolia | ATP synthase beta subunit | 1032 | P:GO:0015986; P:GO:1902600; F:GO:0005524; F:GO:0046933; F:GO:0046961; C:GO:0009535; C:GO:0045261 | P:proton motive force-driven ATP synthesis; P:proton transmembrane transport; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:chloroplast thylakoid membrane; C:proton-transporting ATP synthase complex, catalytic core F(1) | H(+)-exporting diphosphatase; H(+)-transporting two-sector ATPase; Ligases | P:proton motive force-driven ATP synthesis; P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:proton-transporting ATP synthase complex, catalytic core F(1) | view details | ||
| JG650304.1 | Tinospora cordifolia | catalase isozyme 1 | 756 | P:GO:0042542; P:GO:0042744; P:GO:0098869; F:GO:0004096; F:GO:0020037; F:GO:0046872; C:GO:0005777; C:GO:0005886 | P:response to hydrogen peroxide; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:catalase activity; F:heme binding; F:metal ion binding; C:peroxisome; C:plasma membrane | Catalase | P:response to oxidative stress; F:catalase activity; F:heme binding | view details | ||
| JG647909.1 | Tinospora cordifolia | L-ascorbate oxidase homolog | 888 | F:GO:0005507; F:GO:0008447; C:GO:0016021 | F:copper ion binding; F:L-ascorbate oxidase activity; C:integral component of membrane | L-ascorbate oxidase | F:copper ion binding | view details | ||
| JG648385.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 790 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG647759.1 | Tinospora cordifolia | L-ascorbate peroxidase, cytosolic | 776 | P:GO:0000302; P:GO:0034599; P:GO:0042744; P:GO:0098869; F:GO:0016688; F:GO:0020037; F:GO:0046872; C:GO:0009507 | P:response to reactive oxygen species; P:cellular response to oxidative stress; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:L-ascorbate peroxidase activity; F:heme binding; F:metal ion binding; C:chloroplast | L-ascorbate peroxidase | P:response to oxidative stress; P:cellular response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG646590.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 781 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647105.1 | Tinospora cordifolia | serine hydroxymethyltransferase 4 | 819 | P:GO:0006565; P:GO:0019264; P:GO:0032259; P:GO:0035999; P:GO:0046655; F:GO:0004372; F:GO:0008168; F:GO:0008270; F:GO:0030170; F:GO:0070905; C:GO:0005737 | P:L-serine catabolic process; P:glycine biosynthetic process from serine; P:methylation; P:tetrahydrofolate interconversion; P:folic acid metabolic process; F:glycine hydroxymethyltransferase activity; F:methyltransferase activity; F:zinc ion binding; F:pyridoxal phosphate binding; F:serine binding; C:cytoplasm | Glycine hydroxymethyltransferase; Transferring one-carbon groups | P:glycine biosynthetic process from serine; P:tetrahydrofolate interconversion; F:catalytic activity; F:glycine hydroxymethyltransferase activity; F:pyridoxal phosphate binding | view details | ||
| JG648575.1 | Tinospora cordifolia | oxalate--CoA ligase-like | 243 | P:GO:0006631; P:GO:0009698; P:GO:0010030; P:GO:0010214; P:GO:0033611; P:GO:0050832; F:GO:0016207; F:GO:0031956; F:GO:0050203; C:GO:0005737 | P:fatty acid metabolic process; P:phenylpropanoid metabolic process; P:positive regulation of seed germination; P:seed coat development; P:oxalate catabolic process; P:defense response to fungus; F:4-coumarate-CoA ligase activity; F:medium-chain fatty acid-CoA ligase activity; F:oxalate-CoA ligase activity; C:cytoplasm | 4-coumarate--CoA ligase; Oxalate--CoA ligase; Medium-chain acyl-CoA ligase | no GO terms | view details | ||
| JG648998.1 | Tinospora cordifolia | Heat shock 70 kDa protein | 132 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | Acting on acid anhydrides | no IPS match | view details | ||
| JG646927.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 239 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG649729.1 | Tinospora cordifolia | Corytuberine synthase | 735 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| MS687592.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 966 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG648146.1 | Tinospora cordifolia | cytochrome P450 734A1-like | 849 | P:GO:0010268; P:GO:0016131; P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0046872; C:GO:0016020 | P:brassinosteroid homeostasis; P:brassinosteroid metabolic process; P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649348.1 | Tinospora cordifolia | basic endochitinase | 794 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650155.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 801 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646636.1 | Tinospora cordifolia | eukaryotic translation initiation factor 3 subunit D-like | 92 | P:GO:0001732; P:GO:0002191; F:GO:0003735; F:GO:0003743; F:GO:0098808; C:GO:0005840; C:GO:0005852; C:GO:0016282; C:GO:0033290 | P:formation of cytoplasmic translation initiation complex; P:cap-dependent translational initiation; F:structural constituent of ribosome; F:translation initiation factor activity; F:mRNA cap binding; C:ribosome; C:eukaryotic translation initiation factor 3 complex; C:eukaryotic 43S preinitiation complex; C:eukaryotic 48S preinitiation complex | no GO terms | view details | |||
| JG650702.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 844 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649029.1 | Tinospora cordifolia | Phosphoenolpyruvate carboxykinase | 316 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding | view details | ||
| JG648180.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like | 738 | F:GO:0016491 | F:oxidoreductase activity | Oxidoreductases | no GO terms | view details | ||
| JG650497.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 806 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646156.1 | Tinospora cordifolia | monooxygenase 2-like isoform X1 | 844 | F:GO:0016491; C:GO:0016020 | F:oxidoreductase activity; C:membrane | Oxidoreductases | F:FAD binding | view details | ||
| MS687903.1 | Tinospora cordifolia | S-norcoclaurine synthase 1-like | 903 | F:GO:0016491; F:GO:0016829; F:GO:0046872 | F:oxidoreductase activity; F:lyase activity; F:metal ion binding | Oxidoreductases; Lyases | no GO terms | view details | ||
| JG647147.1 | Tinospora cordifolia | N-terminal acetyltransferase A complex auxiliary subunit NAA15 isoform X1 | 749 | P:GO:0017196; F:GO:0004596; C:GO:0005737 | P:N-terminal peptidyl-methionine acetylation; F:peptide alpha-N-acetyltransferase activity; C:cytoplasm | Acyltransferases | no GO terms | view details | ||
| JG647860.1 | Tinospora cordifolia | berberine bridge enzyme-like 26 | 784 | F:GO:0050328; F:GO:0071949; C:GO:0016020 | F:tetrahydroberberine oxidase activity; F:FAD binding; C:membrane | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG645870.1 | Tinospora cordifolia | CLP protease regulatory subunit CLPX1, mitochondrial-like | 846 | P:GO:0006457; P:GO:0006508; P:GO:0030163; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0051082; C:GO:0005759 | P:protein folding; P:proteolysis; P:protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:unfolded protein binding; C:mitochondrial matrix | Acting on acid anhydrides; Acting on peptide bonds (peptidases) | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG648541.1 | Tinospora cordifolia | aspartic proteinase-like | 809 | P:GO:0006508; P:GO:0006629; F:GO:0004190 | P:proteolysis; P:lipid metabolic process; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG647518.1 | Tinospora cordifolia | S-type anion channel SLAH2-like | 816 | P:GO:0006873; P:GO:0015698; P:GO:0034220; F:GO:0008308; C:GO:0005886; C:GO:0016021 | P:cellular ion homeostasis; P:inorganic anion transport; P:ion transmembrane transport; F:voltage-gated anion channel activity; C:plasma membrane; C:integral component of membrane | Translocases | P:cellular ion homeostasis; P:transmembrane transport; F:voltage-gated anion channel activity; C:integral component of membrane | view details | ||
| JG647259.1 | Tinospora cordifolia | MATH domain-containing protein At5g43560 isoform X1 | 741 | no GO terms | view details | |||||
| JG648471.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 753 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648365.1 | Tinospora cordifolia | E3 ubiquitin-protein ligase ORTHRUS 2-like | 822 | P:GO:0006325; P:GO:0010216; P:GO:0016567; F:GO:0003677; F:GO:0046872; F:GO:0061630; C:GO:0005634 | P:chromatin organization; P:maintenance of DNA methylation; P:protein ubiquitination; F:DNA binding; F:metal ion binding; F:ubiquitin protein ligase activity; C:nucleus | Transferases | no GO terms | view details | ||
| JG649534.1 | Tinospora cordifolia | basic endochitinase-like | 808 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650127.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At4g02750-like isoform X2 | 226 | P:GO:0009451; F:GO:0003723; C:GO:0043231 | P:RNA modification; F:RNA binding; C:intracellular membrane-bounded organelle | F:protein binding | view details | |||
| JG646805.1 | Tinospora cordifolia | basic endochitinase | 826 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650507.1 | Tinospora cordifolia | serine decarboxylase-like | 667 | P:GO:0019752; P:GO:1901566; F:GO:0016831; F:GO:0030170 | P:carboxylic acid metabolic process; P:organonitrogen compound biosynthetic process; F:carboxy-lyase activity; F:pyridoxal phosphate binding | Carbon-carbon lyases | F:catalytic activity | view details | ||
| JG647951.1 | Tinospora cordifolia | V-type proton ATPase subunit B 2 | 798 | P:GO:0046034; P:GO:1902600; F:GO:0005524; F:GO:0046961; C:GO:0009507; C:GO:0031090; C:GO:0033180 | P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding; F:proton-transporting ATPase activity, rotational mechanism; C:chloroplast; C:organelle membrane; C:proton-transporting V-type ATPase, V1 domain | H(+)-exporting diphosphatase | P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding | view details | ||
| JG650896.1 | Tinospora cordifolia | cysteine synthase isoform X1 | 831 | P:GO:0006535; F:GO:0004124; C:GO:0005737 | P:cysteine biosynthetic process from serine; F:cysteine synthase activity; C:cytoplasm | Cysteine synthase | P:cysteine biosynthetic process from serine; F:cysteine synthase activity | view details | ||
| JG648452.1 | Tinospora cordifolia | basic endochitinase-like | 822 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649620.1 | Tinospora cordifolia | monodehydroascorbate reductase | 748 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG651005.1 | Tinospora cordifolia | Drug/metabolite transporter | 799 | P:GO:0015860; P:GO:1904823; F:GO:0005345; F:GO:0015211; C:GO:0016021 | P:purine nucleoside transmembrane transport; P:purine nucleobase transmembrane transport; F:purine nucleobase transmembrane transporter activity; F:purine nucleoside transmembrane transporter activity; C:integral component of membrane | Translocases | F:purine nucleoside transmembrane transporter activity; C:integral component of membrane | view details | ||
| JG647898.1 | Tinospora cordifolia | phenylalanine N-monooxygenase-like | 829 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649979.1 | Tinospora cordifolia | Alcohol dehydrogenase class-3 | 263 | P:GO:0006069; P:GO:0008219; P:GO:0010286; P:GO:0046294; P:GO:0048316; F:GO:0004024; F:GO:0008270; F:GO:0080007; F:GO:0106321; F:GO:0106322; C:GO:0005829 | P:ethanol oxidation; P:cell death; P:heat acclimation; P:formaldehyde catabolic process; P:seed development; F:alcohol dehydrogenase activity, zinc-dependent; F:zinc ion binding; F:S-nitrosoglutathione reductase activity; F:S-(hydroxymethyl)glutathione dehydrogenase NADP activity; F:S-(hydroxymethyl)glutathione dehydrogenase NAD activity; C:cytosol | Acting on NADH or NADPH; S-(hydroxymethyl)glutathione dehydrogenase; Alcohol dehydrogenase; Alcohol dehydrogenase (NAD(P)(+)) | no IPS match | view details | ||
| JG648098.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 780 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647961.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 355 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648519.1 | Tinospora cordifolia | methylecgonone reductase | 777 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG648634.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 855 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG647505.1 | Tinospora cordifolia | EEF1A lysine methyltransferase 2-like | 749 | P:GO:0018026; P:GO:0018027; F:GO:0016279; C:GO:0005737 | P:peptidyl-lysine monomethylation; P:peptidyl-lysine dimethylation; F:protein-lysine N-methyltransferase activity; C:cytoplasm | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG646361.1 | Tinospora cordifolia | pre-mRNA-processing-splicing factor 8A | 828 | P:GO:0000244; P:GO:0006508; F:GO:0017070; F:GO:0030619; F:GO:0030620; F:GO:0030623; F:GO:0070122; F:GO:0097157; F:GO:0140492; C:GO:0005682; C:GO:0071013 | P:spliceosomal tri-snRNP complex assembly; P:proteolysis; F:U6 snRNA binding; F:U1 snRNA binding; F:U2 snRNA binding; F:U5 snRNA binding; F:obsolete isopeptidase activity; F:pre-mRNA intronic binding; F:metal-dependent deubiquitinase activity; C:U5 snRNP; C:catalytic step 2 spliceosome | Acting on peptide bonds (peptidases) | P:mRNA splicing, via spliceosome; F:protein binding; F:obsolete isopeptidase activity; F:metal-dependent deubiquitinase activity; C:spliceosomal complex | view details | ||
| JG646219.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 611 | F:GO:0016491; F:GO:0071949; C:GO:0016020 | F:oxidoreductase activity; F:FAD binding; C:membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647711.1 | Tinospora cordifolia | pyrophosphate-energized vacuolar membrane proton pump | 821 | P:GO:1902600; F:GO:0004427; F:GO:0009678; C:GO:0016021 | P:proton transmembrane transport; F:inorganic diphosphatase activity; F:pyrophosphate hydrolysis-driven proton transmembrane transporter activity; C:integral component of membrane | H(+)-exporting diphosphatase; Inorganic diphosphatase | P:proton transmembrane transport; F:inorganic diphosphatase activity; F:pyrophosphate hydrolysis-driven proton transmembrane transporter activity; C:membrane | view details | ||
| JG648994.1 | Tinospora cordifolia | chaperone protein dnaJ 6-like isoform X1 | 126 | no IPS match | view details | |||||
| JG646021.1 | Tinospora cordifolia | ubiquitin carboxyl-terminal hydrolase 12 isoform X6 | 791 | P:GO:0006511; P:GO:0016579; P:GO:0031647; F:GO:0004197; F:GO:0004843; C:GO:0005634; C:GO:0005829 | P:ubiquitin-dependent protein catabolic process; P:protein deubiquitination; P:regulation of protein stability; F:cysteine-type endopeptidase activity; F:cysteine-type deubiquitinase activity; C:nucleus; C:cytosol | Acting on peptide bonds (peptidases); Ubiquitinyl hydrolase 1 | no GO terms | view details | ||
| JG647624.1 | Tinospora cordifolia | heat stress transcription factor A-4b-like | 858 | P:GO:0006357; P:GO:0034605; F:GO:0000978; F:GO:0003700; F:GO:0016740; C:GO:0005634 | P:regulation of transcription by RNA polymerase II; P:cellular response to heat; F:RNA polymerase II cis-regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity; F:transferase activity; C:nucleus | Transferases | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding | view details | ||
| JG646342.1 | Tinospora cordifolia | sucrose nonfermenting 4-like protein isoform X1 | 730 | P:GO:0006468; P:GO:0042149; P:GO:0050790; F:GO:0016208; F:GO:0019887; F:GO:0019901; C:GO:0005634; C:GO:0005737; C:GO:0031588 | P:protein phosphorylation; P:cellular response to glucose starvation; P:regulation of catalytic activity; F:AMP binding; F:protein kinase regulator activity; F:protein kinase binding; C:nucleus; C:cytoplasm; C:nucleotide-activated protein kinase complex | no GO terms | view details | |||
| JG646690.1 | Tinospora cordifolia | berberine bridge enzyme-like 18 | 812 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648629.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 870 | P:GO:0005975; P:GO:0006952; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650428.1 | Tinospora cordifolia | probable protein phosphatase 2C 33 | 713 | P:GO:0006470; F:GO:0017018; C:GO:0005634; C:GO:0005829 | P:protein dephosphorylation; F:myosin phosphatase activity; C:nucleus; C:cytosol | Protein-serine/threonine phosphatase | F:protein serine/threonine phosphatase activity | view details | ||
| JG648191.1 | Tinospora cordifolia | ubiquitin family protein | 822 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG646445.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 837 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG647883.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 803 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG650043.1 | Tinospora cordifolia | strigolactone esterase D14 | 736 | no GO terms | view details | |||||
| JG646769.1 | Tinospora cordifolia | polygalacturonase-inhibiting protein | 789 | C:GO:0110165 | C:cellular anatomical entity | F:protein binding | view details | |||
| JG649439.1 | Tinospora cordifolia | glutathione hydrolase 1-like | 816 | P:GO:0006508; P:GO:0006751; F:GO:0016746; F:GO:0036374; C:GO:0005886; C:GO:0016021 | P:proteolysis; P:glutathione catabolic process; F:acyltransferase activity; F:glutathione hydrolase activity; C:plasma membrane; C:integral component of membrane | Acyltransferases; Glutathione gamma-glutamate hydrolase | P:glutathione catabolic process; F:glutathione hydrolase activity | view details | ||
| JG650604.1 | Tinospora cordifolia | adenosylhomocysteinase 1 | 836 | P:GO:0006730; P:GO:0033353; F:GO:0004013; C:GO:0005829 | P:one-carbon metabolic process; P:S-adenosylmethionine cycle; F:adenosylhomocysteinase activity; C:cytosol | Adenosylhomocysteinase | no GO terms | view details | ||
| JG648917.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 806 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648717.1 | Tinospora cordifolia | actin | 740 | F:GO:0005524; C:GO:0005737; C:GO:0005856 | F:ATP binding; C:cytoplasm; C:cytoskeleton | no GO terms | view details | |||
| JG650866.1 | Tinospora cordifolia | 2-hydroxyacyl-CoA lyase | 805 | P:GO:0001561; F:GO:0000287; F:GO:0016829; F:GO:0030976; C:GO:0005777 | P:fatty acid alpha-oxidation; F:magnesium ion binding; F:lyase activity; F:thiamine pyrophosphate binding; C:peroxisome | Lyases | F:magnesium ion binding; F:thiamine pyrophosphate binding | view details | ||
| JG646040.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 811 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650036.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At4g02750 isoform X2 | 219 | F:GO:0003677; F:GO:0046982; C:GO:0000786 | F:DNA binding; F:protein heterodimerization activity; C:nucleosome | F:protein binding | view details | |||
| JG649383.1 | Tinospora cordifolia | prolyl endopeptidase-like | 842 | P:GO:0006508; F:GO:0004252; F:GO:0070012; C:GO:0005829 | P:proteolysis; F:serine-type endopeptidase activity; F:oligopeptidase activity; C:cytosol | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type endopeptidase activity; F:serine-type peptidase activity | view details | ||
| JG649853.1 | Tinospora cordifolia | Papaver somniferum O-methyltransferase 1 | 763 | P:GO:0009708; P:GO:0032259; F:GO:0008171; F:GO:0030777; F:GO:0046983 | P:benzyl isoquinoline alkaloid biosynthetic process; P:methylation; F:O-methyltransferase activity; F:(S)-scoulerine 9-O-methyltransferase activity; F:protein dimerization activity | (S)-scoulerine 9-O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG650757.1 | Tinospora cordifolia | 4-coumarate--CoA ligase-like 5 | 868 | P:GO:0009611; P:GO:0009617; P:GO:0009695; P:GO:0009698; P:GO:0009753; F:GO:0016207; C:GO:0005777; C:GO:0016021 | P:response to wounding; P:response to bacterium; P:jasmonic acid biosynthetic process; P:phenylpropanoid metabolic process; P:response to jasmonic acid; F:4-coumarate-CoA ligase activity; C:peroxisome; C:integral component of membrane | 4-coumarate--CoA ligase | no GO terms | view details | ||
| JG646739.1 | Tinospora cordifolia | basic endochitinase | 709 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG645615.1 | Tinospora cordifolia | beta-glucosidase 12-like | 814 | P:GO:0005975; P:GO:0009651; P:GO:0019762; F:GO:0008422 | P:carbohydrate metabolic process; P:response to salt stress; P:glucosinolate catabolic process; F:beta-glucosidase activity | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650251.1 | Tinospora cordifolia | polyubiquitin isoform X1 | 795 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG650116.1 | Tinospora cordifolia | Nadp-dependent glyceraldehyde-3-phosphate dehydrogenase | 754 | F:GO:0008911 | F:lactaldehyde dehydrogenase activity | Lactaldehyde dehydrogenase; Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG648840.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 824 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:flavin adenine dinucleotide binding | view details | ||
| JG646703.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 812 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG646968.1 | Tinospora cordifolia | Cannabidiolic acid synthase-like | 335 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:flavin adenine dinucleotide binding | view details | ||
| JG649345.1 | Tinospora cordifolia | myosin-6 isoform X3 | 711 | P:GO:0007015; P:GO:0030050; F:GO:0000146; F:GO:0005516; F:GO:0005524; F:GO:0051015; C:GO:0005737; C:GO:0016459; C:GO:0031982 | P:actin filament organization; P:vesicle transport along actin filament; F:microfilament motor activity; F:calmodulin binding; F:ATP binding; F:actin filament binding; C:cytoplasm; C:myosin complex; C:vesicle | Myosin ATPase | no GO terms | view details | ||
| JG649974.1 | Tinospora cordifolia | Thaumatin-like protein | 181 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG647222.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 760 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG649507.1 | Tinospora cordifolia | Neutral/alkaline nonlysosomal ceramidase | 871 | P:GO:0046514; F:GO:0017040; F:GO:0102121; C:GO:0016021 | P:ceramide catabolic process; F:N-acylsphingosine amidohydrolase activity; F:ceramidase activity; C:integral component of membrane | Ceramidase | P:ceramide catabolic process; F:N-acylsphingosine amidohydrolase activity | view details | ||
| JG647719.1 | Tinospora cordifolia | NAC domain-containing protein 72-like isoform X2 | 836 | P:GO:0006355; F:GO:0003677 | P:regulation of transcription, DNA-templated; F:DNA binding | P:regulation of transcription, DNA-templated; F:DNA binding | view details | |||
| JG646390.1 | Tinospora cordifolia | aldehyde dehydrogenase family 2 member B7, mitochondrial-like | 790 | F:GO:0004029; F:GO:0043878 | F:aldehyde dehydrogenase (NAD+) activity; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity | Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG649155.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 834 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG645755.1 | Tinospora cordifolia | arginine--tRNA ligase, chloroplastic/mitochondrial-like isoform X2 | 681 | P:GO:0006420; F:GO:0004814; F:GO:0005524; C:GO:0005737 | P:arginyl-tRNA aminoacylation; F:arginine-tRNA ligase activity; F:ATP binding; C:cytoplasm | Arginine--tRNA ligase | P:arginyl-tRNA aminoacylation; F:arginine-tRNA ligase activity; F:ATP binding | view details | ||
| JG649399.1 | Tinospora cordifolia | serine/threonine receptor-like kinase NFP | 754 | P:GO:0016310; F:GO:0016301; C:GO:0016020 | P:phosphorylation; F:kinase activity; C:membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity | view details | ||
| JG650854.1 | Tinospora cordifolia | protein MEI2-like 2 | 724 | F:GO:0003723 | F:RNA binding | no GO terms | view details | |||
| JG650142.1 | Tinospora cordifolia | methionine s-methyltransferase | 826 | P:GO:0001887; P:GO:0032259; P:GO:0046500; P:GO:1901566; F:GO:0008483; F:GO:0030170; F:GO:0030732 | P:selenium compound metabolic process; P:methylation; P:S-adenosylmethionine metabolic process; P:organonitrogen compound biosynthetic process; F:transaminase activity; F:pyridoxal phosphate binding; F:methionine S-methyltransferase activity | Transferring nitrogenous groups; Methionine S-methyltransferase | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG646801.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 744 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646779.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 864 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG649124.1 | Tinospora cordifolia | polyubiquitin 10 | 866 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | F:protein binding | view details | |||
| JG650592.1 | Tinospora cordifolia | O-fucosyltransferase 16-like | 837 | P:GO:0006004; F:GO:0016757; C:GO:0016021 | P:fucose metabolic process; F:glycosyltransferase activity; C:integral component of membrane | Glycosyltransferases | no GO terms | view details | ||
| JG650501.1 | Tinospora cordifolia | actin-like | 809 | F:GO:0005524; C:GO:0005737; C:GO:0005856 | F:ATP binding; C:cytoplasm; C:cytoskeleton | no GO terms | view details | |||
| LY749102.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1617 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650993.1 | Tinospora cordifolia | basic endochitinase | 755 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648907.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 789 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| MS687779.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 1596 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648209.1 | Tinospora cordifolia | cell division cycle protein 123 homolog | 734 | P:GO:0051301; C:GO:0005737 | P:cell division; C:cytoplasm | no GO terms | view details | |||
| JG650656.1 | Tinospora cordifolia | Cyclin-dependent kinase f-4 | 876 | P:GO:0006468; P:GO:0035556; F:GO:0004674; F:GO:0005524; C:GO:0005634; C:GO:0005737; C:GO:0016021 | P:protein phosphorylation; P:intracellular signal transduction; F:protein serine/threonine kinase activity; F:ATP binding; C:nucleus; C:cytoplasm; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG649721.1 | Tinospora cordifolia | ---NA--- | 92 | no IPS match | view details | |||||
| JG648135.1 | Tinospora cordifolia | Enolase | 793 | P:GO:0006096; F:GO:0000287; F:GO:0004634; C:GO:0000015 | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | Phosphopyruvate hydratase | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | view details | ||
| LQ667397.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 1596 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650875.1 | Tinospora cordifolia | Cytochrome p450 | 784 | P:GO:0019438; P:GO:1901362; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646409.1 | Tinospora cordifolia | chalcone synthase | 801 | P:GO:0030639; F:GO:0016210; F:GO:0102128 | P:polyketide biosynthetic process; F:naringenin-chalcone synthase activity; F:chalcone synthase activity | Chalcone synthase | P:biosynthetic process; F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG648723.1 | Tinospora cordifolia | basic endochitinase-like | 818 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647658.1 | Tinospora cordifolia | threonylcarbamoyladenosine tRNA methylthiotransferase | 810 | P:GO:0035600; F:GO:0035598; F:GO:0046872; F:GO:0051539; F:GO:0061712; C:GO:0005783; C:GO:0016021 | P:tRNA methylthiolation; F:N6-threonylcarbomyladenosine methylthiotransferase activity; F:metal ion binding; F:4 iron, 4 sulfur cluster binding; F:tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase; C:endoplasmic reticulum; C:integral component of membrane | tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase | F:catalytic activity; F:methylthiotransferase activity; F:iron-sulfur cluster binding; F:4 iron, 4 sulfur cluster binding | view details | ||
| JG649023.1 | Tinospora cordifolia | peroxisomal fatty acid beta-oxidation multifunctional protein AIM1 | 252 | P:GO:0006635; F:GO:0003857; F:GO:0004165; F:GO:0004300; F:GO:0008692; F:GO:0070403; C:GO:0005777; C:GO:0016021 | P:fatty acid beta-oxidation; F:3-hydroxyacyl-CoA dehydrogenase activity; F:delta(3)-delta(2)-enoyl-CoA isomerase activity; F:enoyl-CoA hydratase activity; F:3-hydroxybutyryl-CoA epimerase activity; F:NAD+ binding; C:peroxisome; C:integral component of membrane | 3-hydroxyacyl-CoA dehydrogenase; Delta(3)-Delta(2)-enoyl-CoA isomerase; Enoyl-CoA hydratase; 3-hydroxybutyryl-CoA epimerase | F:catalytic activity | view details | ||
| JG646151.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 761 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG647732.1 | Tinospora cordifolia | proteasome subunit beta type-5 | 795 | P:GO:0010498; F:GO:0004298; C:GO:0005634; C:GO:0005737; C:GO:0019774 | P:proteasomal protein catabolic process; F:threonine-type endopeptidase activity; C:nucleus; C:cytoplasm; C:proteasome core complex, beta-subunit complex | Acting on peptide bonds (peptidases) | P:proteolysis involved in protein catabolic process; F:threonine-type endopeptidase activity; C:proteasome core complex | view details | ||
| MS687902.1 | Tinospora cordifolia | S-norcoclaurine synthase 1-like | 1050 | F:GO:0016491; F:GO:0016829 | F:oxidoreductase activity; F:lyase activity | Oxidoreductases; Lyases | no GO terms | view details | ||
| JG647074.1 | Tinospora cordifolia | probable beta-D-xylosidase 2 | 780 | P:GO:0031222; P:GO:0045493; F:GO:0009044; F:GO:0046556; C:GO:0005576 | P:arabinan catabolic process; P:xylan catabolic process; F:xylan 1,4-beta-xylosidase activity; F:alpha-L-arabinofuranosidase activity; C:extracellular region | Non-reducing end alpha-L-arabinofuranosidase; Xylan 1,4-beta-xylosidase | P:carbohydrate metabolic process; P:xylan catabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:xylan 1,4-beta-xylosidase activity | view details | ||
| JG649000.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 203 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646917.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 739 | no GO terms | view details | |||||
| JG646749.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 819 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650460.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 789 | no GO terms | view details | |||||
| JG648105.1 | Tinospora cordifolia | actin-like | 820 | F:GO:0005524; C:GO:0005737; C:GO:0005856 | F:ATP binding; C:cytoplasm; C:cytoskeleton | no GO terms | view details | |||
| JG648977.1 | Tinospora cordifolia | 40S ribosomal protein S6 | 318 | P:GO:0006412; F:GO:0003735; C:GO:0005737; C:GO:0005840 | P:translation; F:structural constituent of ribosome; C:cytoplasm; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646676.1 | Tinospora cordifolia | basic endochitinase | 792 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650294.1 | Tinospora cordifolia | GTP-binding protein | 744 | P:GO:0006414; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG648273.1 | Tinospora cordifolia | (-)-germacrene D synthase-like | 396 | F:GO:0003824 | F:catalytic activity | F:terpene synthase activity; F:lyase activity | view details | |||
| JG650595.1 | Tinospora cordifolia | ferredoxin--nitrite reductase, chloroplastic-like | 758 | F:GO:0020037; F:GO:0046872; F:GO:0048307; F:GO:0051539; C:GO:0009570 | F:heme binding; F:metal ion binding; F:ferredoxin-nitrite reductase activity; F:4 iron, 4 sulfur cluster binding; C:chloroplast stroma | Ferredoxin--nitrite reductase | F:oxidoreductase activity | view details | ||
| JG650999.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 812 | P:GO:0009733; F:GO:0016881; C:GO:0005737; C:GO:0016021 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm; C:integral component of membrane | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649994.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 799 | P:GO:0005975; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645553.1 | Tinospora cordifolia | putative E3 ubiquitin-protein ligase XBAT31 | 703 | P:GO:0009408; P:GO:0036211; F:GO:0016740; F:GO:0140096 | P:response to heat; P:protein modification process; F:transferase activity; F:catalytic activity, acting on a protein | Transferases | F:protein binding | view details | ||
| JG650727.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 761 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647454.1 | Tinospora cordifolia | pectinesterase/pectinesterase inhibitor PPE8B | 572 | P:GO:0042545; P:GO:0043086; P:GO:0045490; F:GO:0030599; F:GO:0045330; F:GO:0046910; C:GO:0005576 | P:cell wall modification; P:negative regulation of catalytic activity; P:pectin catabolic process; F:pectinesterase activity; F:aspartyl esterase activity; F:pectinesterase inhibitor activity; C:extracellular region | Pectinesterase | P:cell wall modification; F:pectinesterase activity | view details | ||
| JG649710.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 483 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648156.1 | Tinospora cordifolia | polyubiquitin isoform X1 | 750 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG645856.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 854 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG650540.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 752 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648322.1 | Tinospora cordifolia | basic endochitinase-like | 799 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650464.1 | Tinospora cordifolia | alpha-L-arabinofuranosidase 1 | 728 | P:GO:0046373; F:GO:0046556 | P:L-arabinose metabolic process; F:alpha-L-arabinofuranosidase activity | Non-reducing end alpha-L-arabinofuranosidase | no GO terms | view details | ||
| JG650953.1 | Tinospora cordifolia | S-adenosylmethionine synthase 1 | 880 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG647127.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 791 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650469.1 | Tinospora cordifolia | DExH-box ATP-dependent RNA helicase DExH9 isoform X2 | 779 | P:GO:0006401; F:GO:0003723; F:GO:0003724; F:GO:0005524; F:GO:0016787 | P:RNA catabolic process; F:RNA binding; F:RNA helicase activity; F:ATP binding; F:hydrolase activity | RNA helicase | no GO terms | view details | ||
| JG647232.1 | Tinospora cordifolia | elongation factor 1-alpha-like protein | 764 | P:GO:0006414; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG648920.1 | Tinospora cordifolia | glutathione S-transferase L3 | 699 | P:GO:0006749; P:GO:0010731; P:GO:0098869; F:GO:0004364; F:GO:0045174; C:GO:0005737 | P:glutathione metabolic process; P:protein glutathionylation; P:cellular oxidant detoxification; F:glutathione transferase activity; F:glutathione dehydrogenase (ascorbate) activity; C:cytoplasm | Glutathione transferase; Glutathione dehydrogenase (ascorbate) | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG646134.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 799 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650355.1 | Tinospora cordifolia | basic endochitinase-like | 711 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646708.1 | Tinospora cordifolia | probable nucleoredoxin 1 | 815 | P:GO:0098869; F:GO:0004791 | P:cellular oxidant detoxification; F:thioredoxin-disulfide reductase activity | Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:thioredoxin-disulfide reductase activity | view details | ||
| JG646599.1 | Tinospora cordifolia | basic endochitinase-like | 678 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| LY749004.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 966 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG650558.1 | Tinospora cordifolia | ABC transporter G family member 40 | 804 | P:GO:0055085; F:GO:0005524; F:GO:0140359; C:GO:0016021 | P:transmembrane transport; F:ATP binding; F:ABC-type transporter activity; C:integral component of membrane | Translocases | F:ABC-type transporter activity; C:membrane | view details | ||
| JG648989.1 | Tinospora cordifolia | ---NA--- | 63 | no IPS match | view details | |||||
| JG647891.1 | Tinospora cordifolia | protein DETOXIFICATION 29-like | 805 | P:GO:1990961; F:GO:0015297; F:GO:0042910; C:GO:0005774; C:GO:0016021 | P:xenobiotic detoxification by transmembrane export across the plasma membrane; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:vacuolar membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:membrane | view details | ||
| JG650354.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 827 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647192.1 | Tinospora cordifolia | riboflavin biosynthesis protein PYRR, chloroplastic isoform X2 | 733 | P:GO:0009231; P:GO:0009644; P:GO:0009658; P:GO:0046443; P:GO:1901135; F:GO:0008703; F:GO:0008835; F:GO:0016799; F:GO:0050661; C:GO:0009507 | P:riboflavin biosynthetic process; P:response to high light intensity; P:chloroplast organization; P:FAD metabolic process; P:carbohydrate derivative metabolic process; F:5-amino-6-(5-phosphoribosylamino)uracil reductase activity; F:diaminohydroxyphosphoribosylaminopyrimidine deaminase activity; F:hydrolase activity, hydrolyzing N-glycosyl compounds; F:NADP binding; C:chloroplast | Diaminohydroxyphosphoribosylaminopyrimidine deaminase; Glycosylases; 5-amino-6-(5-phosphoribosylamino)uracil reductase | P:riboflavin biosynthetic process; F:catalytic activity; F:5-amino-6-(5-phosphoribosylamino)uracil reductase activity; F:diaminohydroxyphosphoribosylaminopyrimidine deaminase activity | view details | ||
| JG649294.1 | Tinospora cordifolia | Adenosylhomocysteinase | 752 | P:GO:0006730; P:GO:0033353; F:GO:0004013; C:GO:0005829 | P:one-carbon metabolic process; P:S-adenosylmethionine cycle; F:adenosylhomocysteinase activity; C:cytosol | Adenosylhomocysteinase | no GO terms | view details | ||
| JG649915.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 434 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG647206.1 | Tinospora cordifolia | UPF0496 protein 4-like | 860 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG646677.1 | Tinospora cordifolia | Corytuberine synthase | 773 | P:GO:0009058; P:GO:0071704; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0005783; C:GO:0016020 | P:biosynthetic process; P:organic substance metabolic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:endoplasmic reticulum; C:membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647086.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 810 | P:GO:0005975; P:GO:0006952; F:GO:0004553; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:anchored component of plasma membrane | Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646639.1 | Tinospora cordifolia | citrate synthase, glyoxysomal | 798 | P:GO:0005975; P:GO:0006099; F:GO:0003924; F:GO:0004108; F:GO:0005525; C:GO:0005759 | P:carbohydrate metabolic process; P:tricarboxylic acid cycle; F:GTPase activity; F:citrate (Si)-synthase activity; F:GTP binding; C:mitochondrial matrix | Acting on acid anhydrides; Citrate synthase (unknown stereospecificity) | F:acyltransferase activity, acyl groups converted into alkyl on transfer | view details | ||
| JG649965.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 574 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | no IPS match | view details | |||
| JG648154.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 806 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645771.1 | Tinospora cordifolia | sugar transport protein 13 | 625 | P:GO:0006817; P:GO:0015749; P:GO:0050896; F:GO:0015145; F:GO:0015293; C:GO:0016021 | P:phosphate ion transport; P:monosaccharide transmembrane transport; P:response to stimulus; F:monosaccharide transmembrane transporter activity; F:symporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:carbohydrate transmembrane transporter activity; F:transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG649391.1 | Tinospora cordifolia | protein SABRE isoform X2 | 734 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646046.1 | Tinospora cordifolia | basic endochitinase-like | 806 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649079.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 763 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| MS687904.1 | Tinospora cordifolia | S-norcoclaurine synthase 1-like | 1086 | F:GO:0016829 | F:lyase activity | Lyases | no GO terms | view details | ||
| JG646823.1 | Tinospora cordifolia | Pleiotropic drug resistance protein 1 | 435 | P:GO:0055085; F:GO:0005524; F:GO:0140359; C:GO:0005886; C:GO:0016021 | P:transmembrane transport; F:ATP binding; F:ABC-type transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | C:membrane | view details | ||
| JG649985.1 | Tinospora cordifolia | UTP--glucose-1-phosphate uridylyltransferase | 728 | P:GO:0006011; F:GO:0003983; C:GO:0016021 | P:UDP-glucose metabolic process; F:UTP:glucose-1-phosphate uridylyltransferase activity; C:integral component of membrane | UTP-monosaccharide-1-phosphate uridylyltransferase; UTP--glucose-1-phosphate uridylyltransferase | P:UDP-glucose metabolic process; F:UTP:glucose-1-phosphate uridylyltransferase activity; F:uridylyltransferase activity | view details | ||
| JG648615.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 806 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645836.1 | Tinospora cordifolia | Protein NRT1/ PTR family 5.15 | 503 | P:GO:0006817; P:GO:0006857; P:GO:0050896; P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:phosphate ion transport; P:oligopeptide transport; P:response to stimulus; P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| JG645705.1 | Tinospora cordifolia | stomatal closure-related actin-binding protein 1-like | 694 | P:GO:0007015; P:GO:0010119; F:GO:0003779; C:GO:0016021 | P:actin filament organization; P:regulation of stomatal movement; F:actin binding; C:integral component of membrane | P:actin filament organization; P:regulation of stomatal movement; F:actin binding | view details | |||
| JG647237.1 | Tinospora cordifolia | elongation factor 1-alpha-like protein | 770 | P:GO:0006414; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG649331.1 | Tinospora cordifolia | probable ATP synthase 24 kDa subunit, mitochondrial | 728 | P:GO:0009555 | P:pollen development | P:pollen development | view details | |||
| JG648485.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 762 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG646744.1 | Tinospora cordifolia | dihydrolipoyl dehydrogenase 1, mitochondrial | 799 | P:GO:0045454; F:GO:0004148; F:GO:0050660; C:GO:0005739; C:GO:0045252 | P:cell redox homeostasis; F:dihydrolipoyl dehydrogenase activity; F:flavin adenine dinucleotide binding; C:mitochondrion; C:oxoglutarate dehydrogenase complex | Dihydrolipoyl dehydrogenase | P:cell redox homeostasis; F:oxidoreductase activity | view details | ||
| JG649897.1 | Tinospora cordifolia | Hsp70 family protein | 401 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG649552.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 758 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647333.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 796 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650331.1 | Tinospora cordifolia | polyphenol oxidase I, chloroplastic-like | 822 | P:GO:0046148; F:GO:0004097; F:GO:0004503; F:GO:0046872; C:GO:0016021 | P:pigment biosynthetic process; F:catechol oxidase activity; F:tyrosinase activity; F:metal ion binding; C:integral component of membrane | Tyrosinase; Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649038.1 | Tinospora cordifolia | succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial-like | 541 | P:GO:0006099; P:GO:0006121; F:GO:0008177; F:GO:0009055; F:GO:0050660; C:GO:0005749 | P:tricarboxylic acid cycle; P:mitochondrial electron transport, succinate to ubiquinone; F:succinate dehydrogenase (ubiquinone) activity; F:electron transfer activity; F:flavin adenine dinucleotide binding; C:mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | Succinate dehydrogenase (quinone) | no GO terms | view details | ||
| MS688298.1 | Tinospora cordifolia | RS-norcoclaurine 6-O-methyltransferase-like protein | 1026 | P:GO:0009821; P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757; F:GO:0042802; F:GO:0046983 | P:alkaloid biosynthetic process; P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; F:identical protein binding; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG649555.1 | Tinospora cordifolia | basic endochitinase-like | 823 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647213.1 | Tinospora cordifolia | pectinesterase 2 | 844 | P:GO:0042545; P:GO:0043086; P:GO:0045490; F:GO:0030599; F:GO:0045330; F:GO:0046910; C:GO:0005576 | P:cell wall modification; P:negative regulation of catalytic activity; P:pectin catabolic process; F:pectinesterase activity; F:aspartyl esterase activity; F:pectinesterase inhibitor activity; C:extracellular region | Pectinesterase | P:cell wall modification; F:enzyme inhibitor activity; F:pectinesterase activity | view details | ||
| JG649743.1 | Tinospora cordifolia | laccase-7-like | 672 | F:GO:0016491; F:GO:0046872; C:GO:0005576 | F:oxidoreductase activity; F:metal ion binding; C:extracellular region | Oxidoreductases | no GO terms | view details | ||
| JG646235.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 388 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | no IPS match | view details | ||
| JG650470.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 739 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649963.1 | Tinospora cordifolia | histone H3 | 224 | F:GO:0003677; F:GO:0046982; C:GO:0000786; C:GO:0005634 | F:DNA binding; F:protein heterodimerization activity; C:nucleosome; C:nucleus | no IPS match | view details | |||
| JG647031.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 746 | P:GO:0009058; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649013.1 | Tinospora cordifolia | ADP-ribosylation factor 2-B | 414 | P:GO:0006886; P:GO:0016192; P:GO:0050790; F:GO:0003729; F:GO:0003924; F:GO:0005525; F:GO:0016004; C:GO:0005794 | P:intracellular protein transport; P:vesicle-mediated transport; P:regulation of catalytic activity; F:mRNA binding; F:GTPase activity; F:GTP binding; F:phospholipase activator activity; C:Golgi apparatus | Acting on acid anhydrides | no IPS match | view details | ||
| JG648380.1 | Tinospora cordifolia | basic endochitinase | 827 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646439.1 | Tinospora cordifolia | arogenate dehydrogenase 2, chloroplastic-like | 793 | P:GO:0006571; F:GO:0004665; F:GO:0008977; F:GO:0033730 | P:tyrosine biosynthetic process; F:prephenate dehydrogenase (NADP+) activity; F:prephenate dehydrogenase (NAD+) activity; F:arogenate dehydrogenase (NADP+) activity | Arogenate dehydrogenase (NADP(+)); Prephenate dehydrogenase; Prephenate dehydrogenase (NADP(+)); Arogenate dehydrogenase (NAD(P)(+)) | P:tyrosine biosynthetic process; F:prephenate dehydrogenase (NADP+) activity; F:prephenate dehydrogenase (NAD+) activity; F:arogenate dehydrogenase (NADP+) activity | view details | ||
| JG649418.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 740 | no GO terms | view details | |||||
| JG650742.1 | Tinospora cordifolia | BURP domain protein RD22-like | 835 | no GO terms | view details | |||||
| JG648983.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 454 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650872.1 | Tinospora cordifolia | heat shock 70 kDa protein, mitochondrial | 734 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005739 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:mitochondrion | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG647403.1 | Tinospora cordifolia | Adenosylhomocysteinase | 714 | P:GO:0006730; P:GO:0033353; F:GO:0004013; C:GO:0005829 | P:one-carbon metabolic process; P:S-adenosylmethionine cycle; F:adenosylhomocysteinase activity; C:cytosol | Adenosylhomocysteinase | no GO terms | view details | ||
| JG650597.1 | Tinospora cordifolia | WAT1-related protein At5g64700 | 829 | P:GO:0055085; F:GO:0022857; C:GO:0005886; C:GO:0016021 | P:transmembrane transport; F:transmembrane transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | F:transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG650521.1 | Tinospora cordifolia | Acetate/butyrate--CoA ligase AAE7, peroxisomal | 756 | F:GO:0003987; F:GO:0005524; F:GO:0043759; F:GO:0047760; F:GO:0050218; C:GO:0005829 | F:acetate-CoA ligase activity; F:ATP binding; F:methylbutanoate-CoA ligase activity; F:butyrate-CoA ligase activity; F:propionate-CoA ligase activity; C:cytosol | Medium-chain acyl-CoA ligase; Propionate--CoA ligase | no GO terms | view details | ||
| JG645839.1 | Tinospora cordifolia | cytochrome P450 81Q32-like | 770 | P:GO:0009058; F:GO:0016491; F:GO:0046872 | P:biosynthetic process; F:oxidoreductase activity; F:metal ion binding | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646844.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 570 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647692.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 756 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG646130.1 | Tinospora cordifolia | casein kinase 1-like protein 3 | 836 | P:GO:0006897; P:GO:0007165; P:GO:0018105; F:GO:0004674; F:GO:0005524; C:GO:0005634; C:GO:0005737 | P:endocytosis; P:signal transduction; P:peptidyl-serine phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG648343.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 774 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645948.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 698 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646154.1 | Tinospora cordifolia | basic endochitinase | 860 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648009.1 | Tinospora cordifolia | ---NA--- | 53 | no IPS match | view details | |||||
| JG646610.1 | Tinospora cordifolia | 26S proteasome regulatory subunit 6B homolog | 716 | P:GO:0043161; P:GO:1901800; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0036402; C:GO:0005634; C:GO:0005737; C:GO:0008540 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; P:positive regulation of proteasomal protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:proteasome-activating activity; C:nucleus; C:cytoplasm; C:proteasome regulatory particle, base subcomplex | Acting on acid anhydrides; Acting on peptide bonds (peptidases); Proteasome ATPase | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG645621.1 | Tinospora cordifolia | putative disease resistance protein RGA1 | 748 | P:GO:0006952; F:GO:0043531 | P:defense response; F:ADP binding | P:defense response; F:ADP binding | view details | |||
| JG650204.1 | Tinospora cordifolia | omega-6 fatty acid desaturase, chloroplastic | 832 | P:GO:0006629; C:GO:0016021 | P:lipid metabolic process; C:integral component of membrane | P:lipid metabolic process | view details | |||
| JG650132.1 | Tinospora cordifolia | protein transport protein SEC24 | 773 | P:GO:0006886; P:GO:0010584; P:GO:0048658; P:GO:0090110; F:GO:0000149; F:GO:0008270; C:GO:0030127; C:GO:0070971 | P:intracellular protein transport; P:pollen exine formation; P:anther wall tapetum development; P:COPII-coated vesicle cargo loading; F:SNARE binding; F:zinc ion binding; C:COPII vesicle coat; C:endoplasmic reticulum exit site | P:intracellular protein transport; P:endoplasmic reticulum to Golgi vesicle-mediated transport; F:zinc ion binding; C:COPII vesicle coat | view details | |||
| JG650289.1 | Tinospora cordifolia | cytochrome P450 81E8-like | 814 | P:GO:0009058; F:GO:0005488; F:GO:0016491 | P:biosynthetic process; F:binding; F:oxidoreductase activity | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648186.1 | Tinospora cordifolia | polyubiquitin isoform X1 | 789 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG649712.1 | Tinospora cordifolia | probable glutamyl endopeptidase, chloroplastic isoform X2 | 715 | P:GO:0006508; F:GO:0004252 | P:proteolysis; F:serine-type endopeptidase activity | Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| JG647289.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 661 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG649068.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 828 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG647976.1 | Tinospora cordifolia | zinc protease PQQL-like isoform X1 | 665 | P:GO:0006508; F:GO:0004222; F:GO:0046872 | P:proteolysis; F:metalloendopeptidase activity; F:metal ion binding | Acting on peptide bonds (peptidases) | F:metal ion binding | view details | ||
| JG648610.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 865 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648703.1 | Tinospora cordifolia | basic endochitinase-like | 820 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648900.1 | Tinospora cordifolia | ADP,ATP carrier protein, mitochondrial-like | 789 | P:GO:0140021; P:GO:1990544; F:GO:0005471; C:GO:0005743; C:GO:0016021 | P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane; C:integral component of membrane | Translocases | P:transmembrane transport; P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane | view details | ||
| JG649505.1 | Tinospora cordifolia | monodehydroascorbate reductase | 727 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG646863.1 | Tinospora cordifolia | putative E3 ubiquitin-protein ligase XBAT31 | 615 | P:GO:0009408; P:GO:0036211; F:GO:0016740; F:GO:0140096; C:GO:0016021 | P:response to heat; P:protein modification process; F:transferase activity; F:catalytic activity, acting on a protein; C:integral component of membrane | Transferases | F:protein binding | view details | ||
| JG649901.1 | Tinospora cordifolia | glutathione S-transferase L3-like | 55 | P:GO:0006749; F:GO:0004364; F:GO:0016740; C:GO:0016020; C:GO:0016021 | P:glutathione metabolic process; F:glutathione transferase activity; F:transferase activity; C:membrane; C:integral component of membrane | no IPS match | view details | |||
| JG648679.1 | Tinospora cordifolia | glutamate-1-semialdehyde 2,1-aminomutase, chloroplastic-like | 763 | P:GO:0006782; P:GO:0015995; F:GO:0008483; F:GO:0030170; F:GO:0042286; C:GO:0009507 | P:protoporphyrinogen IX biosynthetic process; P:chlorophyll biosynthetic process; F:transaminase activity; F:pyridoxal phosphate binding; F:glutamate-1-semialdehyde 2,1-aminomutase activity; C:chloroplast | Transferring nitrogenous groups; Glutamate-1-semialdehyde 2,1-aminomutase | F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG649245.1 | Tinospora cordifolia | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | 147 | P:GO:0006120; P:GO:0022900; F:GO:0008137; F:GO:0010181; F:GO:0016491; F:GO:0046872; F:GO:0051287; F:GO:0051536; F:GO:0051539; C:GO:0005739; C:GO:0005743; C:GO:0005747; C:GO:0016020; C:GO:0045271; C:GO:0070469 | P:mitochondrial electron transport, NADH to ubiquinone; P:electron transport chain; F:NADH dehydrogenase (ubiquinone) activity; F:FMN binding; F:oxidoreductase activity; F:metal ion binding; F:NAD binding; F:iron-sulfur cluster binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrion; C:mitochondrial inner membrane; C:mitochondrial respiratory chain complex I; C:membrane; C:respiratory chain complex I; C:respirasome | no IPS match | view details | |||
| JG649757.1 | Tinospora cordifolia | cellulose synthase-like protein e1 | 614 | P:GO:0030244; F:GO:0016760; C:GO:0016021 | P:cellulose biosynthetic process; F:cellulose synthase (UDP-forming) activity; C:integral component of membrane | Cellulose synthase (UDP-forming) | P:cellulose biosynthetic process; F:cellulose synthase (UDP-forming) activity; C:membrane | view details | ||
| JG649322.1 | Tinospora cordifolia | thylakoidal processing peptidase 1, chloroplastic-like | 716 | P:GO:0006465; P:GO:0010027; F:GO:0004252; C:GO:0005887; C:GO:0009535 | P:signal peptide processing; P:thylakoid membrane organization; F:serine-type endopeptidase activity; C:integral component of plasma membrane; C:chloroplast thylakoid membrane | Acting on peptide bonds (peptidases) | P:signal peptide processing; P:proteolysis; F:serine-type endopeptidase activity; F:serine-type peptidase activity; C:membrane | view details | ||
| JG647988.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1 | 843 | F:GO:0016616; F:GO:0016621 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:cinnamoyl-CoA reductase activity | Cinnamoyl-CoA reductase; Acting on the CH-OH group of donors | no GO terms | view details | ||
| JG649751.1 | Tinospora cordifolia | probable pectinesterase/pectinesterase inhibitor 7 | 426 | F:GO:0030599; F:GO:0046910 | F:pectinesterase activity; F:pectinesterase inhibitor activity | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG649805.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 791 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648964.1 | Tinospora cordifolia | ribonuclease 1-like | 513 | P:GO:0006401; P:GO:0090502; F:GO:0003723; F:GO:0033897; C:GO:0005576 | P:RNA catabolic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA binding; F:ribonuclease T2 activity; C:extracellular region | Acting on ester bonds; Ribonuclease T(2) | F:RNA binding; F:ribonuclease T2 activity | view details | ||
| JG645906.1 | Tinospora cordifolia | beta-glucosidase BoGH3B-like | 818 | P:GO:0009251; F:GO:0008422; C:GO:0005576 | P:glucan catabolic process; F:beta-glucosidase activity; C:extracellular region | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650316.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 768 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649934.1 | Tinospora cordifolia | zinc finger A20 and AN1 domain-containing stress-associated protein 6-like | 188 | F:GO:0003677; F:GO:0008270 | F:DNA binding; F:zinc ion binding | F:DNA binding; F:zinc ion binding | view details | |||
| JG646976.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 661 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| MT850045.1 | Tinospora cordifolia | maturase K | 1530 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG649749.1 | Tinospora cordifolia | bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 | 504 | P:GO:0006012; P:GO:0006364; F:GO:0003723; F:GO:0003978; C:GO:0005829 | P:galactose metabolic process; P:rRNA processing; F:RNA binding; F:UDP-glucose 4-epimerase activity; C:cytosol | UDP-glucose 4-epimerase | no GO terms | view details | ||
| JG650655.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 809 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG645633.1 | Tinospora cordifolia | GDSL esterase/lipase 7-like | 770 | F:GO:0016788; C:GO:0016020 | F:hydrolase activity, acting on ester bonds; C:membrane | Acting on ester bonds | F:hydrolase activity, acting on ester bonds | view details | ||
| JG647337.1 | Tinospora cordifolia | probable protein phosphatase 2C 60 | 775 | P:GO:0006470; F:GO:0017018; F:GO:0046872 | P:protein dephosphorylation; F:myosin phosphatase activity; F:metal ion binding | Protein-serine/threonine phosphatase | P:protein dephosphorylation; F:protein serine/threonine phosphatase activity | view details | ||
| JG647538.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 699 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646473.1 | Tinospora cordifolia | lipoxygenase 6, chloroplastic | 798 | P:GO:0006633; P:GO:0009611; P:GO:0009695; P:GO:0031408; P:GO:0034440; F:GO:0016165; F:GO:0046872; C:GO:0005737 | P:fatty acid biosynthetic process; P:response to wounding; P:jasmonic acid biosynthetic process; P:oxylipin biosynthetic process; P:lipid oxidation; F:linoleate 13S-lipoxygenase activity; F:metal ion binding; C:cytoplasm | Linoleate 13S-lipoxygenase | F:oxidoreductase activity; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; F:metal ion binding | view details | ||
| JG650134.1 | Tinospora cordifolia | Strigolactone esterase d14 | 849 | no GO terms | view details | |||||
| JG649012.1 | Tinospora cordifolia | 6-phosphogluconate dehydrogenase, decarboxylating | 312 | P:GO:0009051; P:GO:0046177; F:GO:0004616; F:GO:0050661; C:GO:0005829 | P:pentose-phosphate shunt, oxidative branch; P:D-gluconate catabolic process; F:phosphogluconate dehydrogenase (decarboxylating) activity; F:NADP binding; C:cytosol | Phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | F:phosphogluconate dehydrogenase (decarboxylating) activity; F:NADP binding | view details | ||
| JG645657.1 | Tinospora cordifolia | ---NA--- | 64 | no IPS match | view details | |||||
| JG648582.1 | Tinospora cordifolia | Ribosomal protein L2 | 729 | P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646330.1 | Tinospora cordifolia | CBL-interacting serine/threonine-protein kinase 8-like isoform X2 | 601 | P:GO:0006468; P:GO:0010167; P:GO:0035556; P:GO:0048364; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310 | P:protein phosphorylation; P:response to nitrate; P:intracellular signal transduction; P:root development; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity | Dual-specificity kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG646056.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 796 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646762.1 | Tinospora cordifolia | patatin-like protein 2 | 751 | P:GO:0016042; F:GO:0004620; F:GO:0045735; F:GO:0047372; C:GO:0005773 | P:lipid catabolic process; F:phospholipase activity; F:nutrient reservoir activity; F:acylglycerol lipase activity; C:vacuole | Acylglycerol lipase; Carboxylesterase | P:lipid metabolic process | view details | ||
| MS688405.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein At5g02240-like | 768 | F:GO:0016491 | F:oxidoreductase activity | Oxidoreductases | F:oxidoreductase activity | view details | ||
| JG646581.1 | Tinospora cordifolia | basic endochitinase-like | 798 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650483.1 | Tinospora cordifolia | AAA-ATPase At5g57480-like | 857 | F:GO:0005524; F:GO:0016887 | F:ATP binding; F:ATP hydrolysis activity | Acting on acid anhydrides | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG647903.1 | Tinospora cordifolia | UPF0496 protein 4-like | 816 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG646362.1 | Tinospora cordifolia | basic endochitinase-like | 773 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649770.1 | Tinospora cordifolia | B3 domain-containing transcription repressor VAL2 | 582 | F:GO:0003677; F:GO:0008270; C:GO:0005634 | F:DNA binding; F:zinc ion binding; C:nucleus | no GO terms | view details | |||
| JG649596.1 | Tinospora cordifolia | phosphoenolpyruvate/phosphate translocator 1, chloroplastic-like | 731 | P:GO:0015713; P:GO:0089722; F:GO:0015120; F:GO:0015121; C:GO:0005794; C:GO:0016021; C:GO:0031969 | P:phosphoglycerate transmembrane transport; P:phosphoenolpyruvate transmembrane transport; F:phosphoglycerate transmembrane transporter activity; F:phosphoenolpyruvate:phosphate antiporter activity; C:Golgi apparatus; C:integral component of membrane; C:chloroplast membrane | Translocases | no GO terms | view details | ||
| JG649827.1 | Tinospora cordifolia | polygalacturonase inhibitor | 611 | C:GO:0016020 | C:membrane | F:protein binding | view details | |||
| JG650816.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 830 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| DQ353864.2 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1409 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG648392.1 | Tinospora cordifolia | sulfate transporter 1.3-like | 727 | P:GO:0090502; P:GO:1902358; F:GO:0003676; F:GO:0004523; F:GO:0008271; F:GO:0015301; C:GO:0005887 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; P:sulfate transmembrane transport; F:nucleic acid binding; F:RNA-DNA hybrid ribonuclease activity; F:secondary active sulfate transmembrane transporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane | Acting on ester bonds; Translocases; Ribonuclease H | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG645978.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 829 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG650890.1 | Tinospora cordifolia | cellulose synthase 1B | 799 | P:GO:0030244; P:GO:0071555; F:GO:0016760; F:GO:0046872; C:GO:0005886; C:GO:0016021 | P:cellulose biosynthetic process; P:cell wall organization; F:cellulose synthase (UDP-forming) activity; F:metal ion binding; C:plasma membrane; C:integral component of membrane | Cellulose synthase (UDP-forming) | P:cellulose biosynthetic process; F:cellulose synthase (UDP-forming) activity; C:membrane | view details | ||
| JG646618.1 | Tinospora cordifolia | histone deacetylase 19 | 803 | P:GO:0006325; P:GO:0006357; P:GO:0016567; P:GO:0016575; F:GO:0004407; F:GO:0046872; F:GO:0061630; C:GO:0005634 | P:chromatin organization; P:regulation of transcription by RNA polymerase II; P:protein ubiquitination; P:histone deacetylation; F:histone deacetylase activity; F:metal ion binding; F:ubiquitin protein ligase activity; C:nucleus | Histone deacetylase; Transferases | P:histone deacetylation; F:histone deacetylase activity | view details | ||
| JG648822.1 | Tinospora cordifolia | Pathogenesis-related protein 1 | 397 | P:GO:0009409; P:GO:0009611; P:GO:0009617; P:GO:0009646; P:GO:0009651; P:GO:0009733; P:GO:0009735; P:GO:0009737; P:GO:0009739; P:GO:0009751; P:GO:0042542; P:GO:0046688; P:GO:0050790; P:GO:0050794; P:GO:0050832; F:GO:0003723; F:GO:0004540; C:GO:0005622 | P:response to cold; P:response to wounding; P:response to bacterium; P:response to absence of light; P:response to salt stress; P:response to auxin; P:response to cytokinin; P:response to abscisic acid; P:response to gibberellin; P:response to salicylic acid; P:response to hydrogen peroxide; P:response to copper ion; P:regulation of catalytic activity; P:regulation of cellular process; P:defense response to fungus; F:RNA binding; F:ribonuclease activity; C:intracellular anatomical structure | Acting on ester bonds | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | ||
| JG648084.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 852 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648504.1 | Tinospora cordifolia | 60S ribosomal protein L7-2-like | 818 | P:GO:0000463; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG645894.1 | Tinospora cordifolia | berberine bridge enzyme-like 26 | 724 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646544.1 | Tinospora cordifolia | chaperone protein DnaJ-like | 346 | C:GO:0005634; C:GO:0005737 | C:nucleus; C:cytoplasm | no GO terms | view details | |||
| JG646340.1 | Tinospora cordifolia | UPF0496 protein 4-like | 733 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG650273.1 | Tinospora cordifolia | probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 | 776 | P:GO:0005992; P:GO:0070413; F:GO:0016740 | P:trehalose biosynthetic process; P:trehalose metabolism in response to stress; F:transferase activity | Transferases | P:trehalose biosynthetic process; F:catalytic activity | view details | ||
| JG645739.1 | Tinospora cordifolia | outer envelope protein 61-like | 696 | C:GO:0016021 | C:integral component of membrane | F:protein binding | view details | |||
| JG646460.1 | Tinospora cordifolia | protein ELC-like | 835 | P:GO:0008333; P:GO:0015031; P:GO:0036211; F:GO:0043130; C:GO:0000813 | P:endosome to lysosome transport; P:protein transport; P:protein modification process; F:ubiquitin binding; C:ESCRT I complex | P:protein transport; P:protein modification process | view details | |||
| JG649267.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 718 | no GO terms | view details | |||||
| JG648804.1 | Tinospora cordifolia | basic endochitinase-like | 828 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649713.1 | Tinospora cordifolia | acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X3 | 606 | P:GO:0009698; P:GO:0019427; F:GO:0003987; F:GO:0005524; F:GO:0016207; F:GO:0016208; C:GO:0009536; C:GO:0016021 | P:phenylpropanoid metabolic process; P:acetyl-CoA biosynthetic process from acetate; F:acetate-CoA ligase activity; F:ATP binding; F:4-coumarate-CoA ligase activity; F:AMP binding; C:plastid; C:integral component of membrane | 4-coumarate--CoA ligase | no GO terms | view details | ||
| JG649154.1 | Tinospora cordifolia | salicylic acid-binding protein 2-like | 842 | P:GO:0009694; P:GO:0009696; F:GO:0080030; F:GO:0080031; F:GO:0080032 | P:jasmonic acid metabolic process; P:salicylic acid metabolic process; F:methyl indole-3-acetate esterase activity; F:methyl salicylate esterase activity; F:methyl jasmonate esterase activity | Carboxylesterase | no GO terms | view details | ||
| JG650493.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 786 | P:GO:0005975; P:GO:0006952; F:GO:0004553; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:anchored component of plasma membrane | Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648095.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 816 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG650533.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 829 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649947.1 | Tinospora cordifolia | probable linoleate 9S-lipoxygenase 5 isoform X1 | 381 | P:GO:0006633; P:GO:0031408; P:GO:0034440; F:GO:0016702; F:GO:0046872; C:GO:0005737 | P:fatty acid biosynthetic process; P:oxylipin biosynthetic process; P:lipid oxidation; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; F:metal ion binding; C:cytoplasm | Acting on single donors with incorporation of molecular oxygen (oxygenases). The oxygen incorporated need not be derived from O(2) | F:oxidoreductase activity; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; F:metal ion binding | view details | ||
| JG648125.1 | Tinospora cordifolia | Berberine bridge enzyme-like 26 | 794 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648954.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 793 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG647166.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 796 | P:GO:0005975; P:GO:0006952; F:GO:0004553; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:anchored component of plasma membrane | Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647123.1 | Tinospora cordifolia | cellulose synthase A catalytic subunit 1 [UDP-forming] | 773 | P:GO:0030244; P:GO:0071555; F:GO:0016760; F:GO:0046872; C:GO:0005886; C:GO:0016021 | P:cellulose biosynthetic process; P:cell wall organization; F:cellulose synthase (UDP-forming) activity; F:metal ion binding; C:plasma membrane; C:integral component of membrane | Cellulose synthase (UDP-forming) | P:cellulose biosynthetic process; F:cellulose synthase (UDP-forming) activity; C:membrane | view details | ||
| JG649001.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At5g15340, mitochondrial | 336 | P:GO:0006810; F:GO:0008270 | P:transport; F:zinc ion binding | F:protein binding | view details | |||
| JG649882.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 666 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | no GO terms | view details | ||
| JG648805.1 | Tinospora cordifolia | basic endochitinase-like | 822 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647677.1 | Tinospora cordifolia | alpha-glucan water dikinase 2 isoform X1 | 770 | P:GO:0016310; F:GO:0005524; F:GO:0016301 | P:phosphorylation; F:ATP binding; F:kinase activity | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG646940.1 | Tinospora cordifolia | ATP-dependent 6-phosphofructokinase 5, chloroplastic-like | 245 | P:GO:0006002; P:GO:0061615; F:GO:0003872; F:GO:0005524; F:GO:0046872; C:GO:0005737 | P:fructose 6-phosphate metabolic process; P:glycolytic process through fructose-6-phosphate; F:6-phosphofructokinase activity; F:ATP binding; F:metal ion binding; C:cytoplasm | 6-phosphofructokinase | no IPS match | view details | ||
| LQ667394.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1569 | P:GO:0006259; P:GO:0006974; P:GO:0009058; P:GO:0051276; P:GO:0098542; F:GO:0000166; F:GO:0004386; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:DNA metabolic process; P:cellular response to DNA damage stimulus; P:biosynthetic process; P:chromosome organization; P:defense response to other organism; F:nucleotide binding; F:helicase activity; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649951.1 | Tinospora cordifolia | ADP-ribosylation factor 1 | 479 | P:GO:0006886; P:GO:0016192; F:GO:0003925; F:GO:0005525; C:GO:0005794 | P:intracellular protein transport; P:vesicle-mediated transport; F:G protein activity; F:GTP binding; C:Golgi apparatus | Acting on acid anhydrides; Heterotrimeric G-protein GTPase; Small monomeric GTPase | F:GTPase activity; F:GTP binding | view details | ||
| JG649354.1 | Tinospora cordifolia | FRIGIDA-like protein 3 | 796 | P:GO:0009908; P:GO:0030154 | P:flower development; P:cell differentiation | no GO terms | view details | |||
| JG647115.1 | Tinospora cordifolia | (-)-alpha-pinene synthase-like | 779 | F:GO:0016829; F:GO:0046872 | F:lyase activity; F:metal ion binding | Lyases | F:terpene synthase activity; F:lyase activity | view details | ||
| JG649763.1 | Tinospora cordifolia | Proteasome assembly chaperone 2 | 673 | P:GO:0043248; P:GO:0090502; F:GO:0003676; F:GO:0004523; C:GO:0000502; C:GO:0005634; C:GO:0005829 | P:proteasome assembly; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:nucleic acid binding; F:RNA-DNA hybrid ribonuclease activity; C:proteasome complex; C:nucleus; C:cytosol | Acting on ester bonds; Ribonuclease H | no GO terms | view details | ||
| LY340441.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1617 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649879.1 | Tinospora cordifolia | pathogenesis-related protein 1-like | 547 | P:GO:0006952; P:GO:0009607; C:GO:0005615 | P:defense response; P:response to biotic stimulus; C:extracellular space | no GO terms | view details | |||
| JG646482.1 | Tinospora cordifolia | adenosine deaminase-like protein | 845 | P:GO:0006154; P:GO:0046103; F:GO:0004000 | P:adenosine catabolic process; P:inosine biosynthetic process; F:adenosine deaminase activity | Adenosine deaminase | F:deaminase activity | view details | ||
| JG647125.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 866 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649738.1 | Tinospora cordifolia | Heat shock cognate protein 80 | 609 | P:GO:0006457; P:GO:0034605; P:GO:0050821; F:GO:0005524; F:GO:0016887; F:GO:0051082; C:GO:0005829; C:GO:0005886; C:GO:0032991; C:GO:0048471 | P:protein folding; P:cellular response to heat; P:protein stabilization; F:ATP binding; F:ATP hydrolysis activity; F:unfolded protein binding; C:cytosol; C:plasma membrane; C:protein-containing complex; C:perinuclear region of cytoplasm | Acting on acid anhydrides | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; F:unfolded protein binding | view details | ||
| JG649295.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 709 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649264.1 | Tinospora cordifolia | oxalate--CoA ligase | 744 | P:GO:0006631; P:GO:0009698; P:GO:0010030; P:GO:0010214; P:GO:0033611; P:GO:0050832; F:GO:0016207; F:GO:0031956; F:GO:0050203; C:GO:0005737 | P:fatty acid metabolic process; P:phenylpropanoid metabolic process; P:positive regulation of seed germination; P:seed coat development; P:oxalate catabolic process; P:defense response to fungus; F:4-coumarate-CoA ligase activity; F:medium-chain fatty acid-CoA ligase activity; F:oxalate-CoA ligase activity; C:cytoplasm | 4-coumarate--CoA ligase; Oxalate--CoA ligase; Medium-chain acyl-CoA ligase | no GO terms | view details | ||
| JG645794.1 | Tinospora cordifolia | serine/threonine-protein kinase rio2 | 806 | P:GO:0006468; P:GO:0030490; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0008168; F:GO:0106310; C:GO:0005634; C:GO:0005829; C:GO:0030688 | P:protein phosphorylation; P:maturation of SSU-rRNA; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:methyltransferase activity; F:protein serine kinase activity; C:nucleus; C:cytosol; C:preribosome, small subunit precursor | Dual-specificity kinase; Transferring one-carbon groups | P:protein phosphorylation; F:protein kinase activity; F:protein serine/threonine kinase activity; F:ATP binding | view details | ||
| JG648169.1 | Tinospora cordifolia | cytochrome b5 domain-containing protein RLF | 715 | F:GO:0020037; F:GO:0046872; C:GO:0005829 | F:heme binding; F:metal ion binding; C:cytosol | no GO terms | view details | |||
| JG647916.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 767 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648028.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 846 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG650614.1 | Tinospora cordifolia | myosin-6-like isoform X3 | 739 | P:GO:0007015; P:GO:0030050; P:GO:0048767; F:GO:0000146; F:GO:0005516; F:GO:0005524; F:GO:0051015; C:GO:0005737; C:GO:0016459; C:GO:0031982 | P:actin filament organization; P:vesicle transport along actin filament; P:root hair elongation; F:microfilament motor activity; F:calmodulin binding; F:ATP binding; F:actin filament binding; C:cytoplasm; C:myosin complex; C:vesicle | Myosin ATPase | F:cytoskeletal motor activity; F:ATP binding; C:myosin complex | view details | ||
| JG646026.1 | Tinospora cordifolia | probable beta-D-xylosidase 5 | 813 | P:GO:0005975; F:GO:0004553 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | Glycosylases | P:carbohydrate metabolic process; P:xylan catabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:xylan 1,4-beta-xylosidase activity | view details | ||
| JG648722.1 | Tinospora cordifolia | plant intracellular Ras-group-related LRR protein 5-like | 683 | no GO terms | view details | |||||
| JG648327.1 | Tinospora cordifolia | Udp-glycosyltransferase 92a1 | 715 | F:GO:0008194 | F:UDP-glycosyltransferase activity | Glycosyltransferases | no GO terms | view details | ||
| JG645688.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 798 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648708.1 | Tinospora cordifolia | nucleobase-ascorbate transporter 6-like | 811 | P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| JG650842.1 | Tinospora cordifolia | GDSL esterase/lipase 7-like | 784 | F:GO:0016788; C:GO:0016020 | F:hydrolase activity, acting on ester bonds; C:membrane | Acting on ester bonds | F:hydrolase activity, acting on ester bonds | view details | ||
| JG645760.1 | Tinospora cordifolia | flowering time control protein FPA | 780 | P:GO:0006378; P:GO:0009553; P:GO:0009911; P:GO:0010228; P:GO:0031048; F:GO:0003676; F:GO:0003723; C:GO:0000785; C:GO:0110165 | P:mRNA polyadenylation; P:embryo sac development; P:positive regulation of flower development; P:vegetative to reproductive phase transition of meristem; P:heterochromatin assembly by small RNA; F:nucleic acid binding; F:RNA binding; C:chromatin; C:cellular anatomical entity | no GO terms | view details | |||
| KF432030.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 578 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG650640.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 853 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648838.1 | Tinospora cordifolia | glycosyl hydrolase family protein | 779 | P:GO:0005975; F:GO:0004553; C:GO:0005576 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:extracellular region | Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648782.1 | Tinospora cordifolia | mitochondrial phosphate carrier protein 1, mitochondrial-like | 445 | P:GO:1990547; F:GO:0005315; C:GO:0005743; C:GO:0016021 | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity; C:mitochondrial inner membrane; C:integral component of membrane | Translocases | no IPS match | view details | ||
| JG649918.1 | Tinospora cordifolia | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial | 428 | P:GO:0022900; F:GO:0008137; F:GO:0010181; F:GO:0046872; F:GO:0051287; F:GO:0051539; C:GO:0005743; C:GO:0045271 | P:electron transport chain; F:NADH dehydrogenase (ubiquinone) activity; F:FMN binding; F:metal ion binding; F:NAD binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrial inner membrane; C:respiratory chain complex I | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | no GO terms | view details | ||
| JG651006.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 854 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649602.1 | Tinospora cordifolia | ATP-dependent zinc metalloprotease FTSH 2, chloroplastic | 707 | P:GO:0006508; F:GO:0004176; F:GO:0004222; F:GO:0005524; F:GO:0008270; F:GO:0016887; C:GO:0009526; C:GO:0009534; C:GO:0016021 | P:proteolysis; F:ATP-dependent peptidase activity; F:metalloendopeptidase activity; F:ATP binding; F:zinc ion binding; F:ATP hydrolysis activity; C:plastid envelope; C:chloroplast thylakoid; C:integral component of membrane | Acting on peptide bonds (peptidases); Acting on acid anhydrides | no GO terms | view details | ||
| JG647245.1 | Tinospora cordifolia | dihydrolipoyl dehydrogenase 1, mitochondrial | 840 | P:GO:0045454; F:GO:0004148; F:GO:0050660; C:GO:0005739; C:GO:0045252 | P:cell redox homeostasis; F:dihydrolipoyl dehydrogenase activity; F:flavin adenine dinucleotide binding; C:mitochondrion; C:oxoglutarate dehydrogenase complex | Dihydrolipoyl dehydrogenase | F:oxidoreductase activity | view details | ||
| JG649357.1 | Tinospora cordifolia | eukaryotic translation initiation factor 2D | 784 | P:GO:0001731; F:GO:0003743 | P:formation of translation preinitiation complex; F:translation initiation factor activity | P:formation of translation preinitiation complex; F:RNA binding; F:translation initiation factor activity | view details | |||
| JG648292.1 | Tinospora cordifolia | kinesin-like protein KIN-7D, mitochondrial | 232 | P:GO:0007018; P:GO:0009767; P:GO:0018298; F:GO:0003777; F:GO:0005524; F:GO:0008017; F:GO:0016168; F:GO:0016887; C:GO:0005737; C:GO:0005871; C:GO:0005874; C:GO:0009521; C:GO:0016021 | P:microtubule-based movement; P:photosynthetic electron transport chain; P:obsolete protein-chromophore linkage; F:microtubule motor activity; F:ATP binding; F:microtubule binding; F:chlorophyll binding; F:ATP hydrolysis activity; C:cytoplasm; C:kinesin complex; C:microtubule; C:photosystem; C:integral component of membrane | Acting on acid anhydrides | no IPS match | view details | ||
| JG650471.1 | Tinospora cordifolia | nitrate reductase [NADH] | 802 | P:GO:0006809; P:GO:0042128; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity | Nitrate reductase (NAD(P)H); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG649925.1 | Tinospora cordifolia | lactoylglutathione lyase GLX1-like | 258 | P:GO:0019243; F:GO:0004462; F:GO:0046872; C:GO:0005737 | P:methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione; F:lactoylglutathione lyase activity; F:metal ion binding; C:cytoplasm | Lactoylglutathione lyase | no IPS match | view details | ||
| JG646908.1 | Tinospora cordifolia | Corytuberine synthase | 302 | P:GO:0009820; P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0005783; C:GO:0016020 | P:alkaloid metabolic process; P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:endoplasmic reticulum; C:membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no IPS match | view details | ||
| JG649784.1 | Tinospora cordifolia | mitogen-activated protein kinase kinase kinase NPK1-like isoform X4 | 825 | P:GO:0006468; F:GO:0004672; F:GO:0005524 | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG649108.1 | Tinospora cordifolia | 7-dehydrocholesterol reductase | 631 | P:GO:0016126; P:GO:0016132; F:GO:0009918; F:GO:0016628; C:GO:0030176 | P:sterol biosynthetic process; P:brassinosteroid biosynthetic process; F:sterol delta7 reductase activity; F:oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor; C:integral component of endoplasmic reticulum membrane | Acting on the CH-CH group of donors | P:sterol biosynthetic process; F:oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor; C:membrane | view details | ||
| JG649167.1 | Tinospora cordifolia | basic endochitinase-like | 765 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647357.1 | Tinospora cordifolia | heat shock 70 kDa protein 16-like | 746 | P:GO:0006457; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0005829 | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:cytosol | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG648116.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1 | 788 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | no GO terms | view details | ||
| JG649877.1 | Tinospora cordifolia | 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 | 451 | P:GO:0044237; P:GO:0044238; P:GO:0071704; F:GO:0016616; C:GO:0005783; C:GO:0016020 | P:cellular metabolic process; P:primary metabolic process; P:organic substance metabolic process; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; C:endoplasmic reticulum; C:membrane | Acting on the CH-OH group of donors | P:coenzyme A metabolic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity | view details | ||
| JG648507.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 762 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649435.1 | Tinospora cordifolia | polyphenol oxidase | 763 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648276.1 | Tinospora cordifolia | GTP-binding protein YPTM2 | 455 | F:GO:0003924; F:GO:0005525; C:GO:0009507 | F:GTPase activity; F:GTP binding; C:chloroplast | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG648857.1 | Tinospora cordifolia | elongation factor 1-delta 1-like | 486 | P:GO:0006414; P:GO:0050790; F:GO:0003746; F:GO:0005085; C:GO:0005829; C:GO:0005853 | P:translational elongation; P:regulation of catalytic activity; F:translation elongation factor activity; F:guanyl-nucleotide exchange factor activity; C:cytosol; C:eukaryotic translation elongation factor 1 complex | P:translational elongation; F:translation elongation factor activity | view details | |||
| JG646874.1 | Tinospora cordifolia | 60S ribosomal protein L9 | 496 | P:GO:0002181; F:GO:0003735; F:GO:0019843; C:GO:0022625 | P:cytoplasmic translation; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:ribosome | view details | |||
| MN602448.1 | Tinospora cordifolia | cytochrome c biogenesis protein | 333 | P:GO:0017004; F:GO:0020037; C:GO:0009535; C:GO:0016021 | P:cytochrome complex assembly; F:heme binding; C:chloroplast thylakoid membrane; C:integral component of membrane | P:cytochrome complex assembly; F:heme binding | view details | |||
| MA258231.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1617 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650287.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 839 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647959.1 | Tinospora cordifolia | 40S ribosomal protein S6 | 725 | P:GO:0006412; F:GO:0003735; C:GO:0005737; C:GO:0005840 | P:translation; F:structural constituent of ribosome; C:cytoplasm; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648126.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 767 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650463.1 | Tinospora cordifolia | protein TRANSPORT INHIBITOR RESPONSE 1-like | 773 | P:GO:0009734; P:GO:0031146; C:GO:0005634; C:GO:0019005 | P:auxin-activated signaling pathway; P:SCF-dependent proteasomal ubiquitin-dependent protein catabolic process; C:nucleus; C:SCF ubiquitin ligase complex | no GO terms | view details | |||
| JG647030.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 863 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648993.1 | Tinospora cordifolia | Polygalacturonase inhibitor | 332 | F:GO:0016491; F:GO:0016740 | F:oxidoreductase activity; F:transferase activity | Oxidoreductases; Transferases | F:protein binding | view details | ||
| JG645690.1 | Tinospora cordifolia | polyubiquitin 10 | 841 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | F:protein binding | view details | |||
| JG650336.1 | Tinospora cordifolia | NAC domain-containing protein 72 | 807 | P:GO:0006355; F:GO:0003677 | P:regulation of transcription, DNA-templated; F:DNA binding | P:regulation of transcription, DNA-templated; F:DNA binding | view details | |||
| JG650575.1 | Tinospora cordifolia | caffeic acid O-methyltransferase | 783 | P:GO:0009809; P:GO:0032259; F:GO:0008757; F:GO:0046983; F:GO:0047763 | P:lignin biosynthetic process; P:methylation; F:S-adenosylmethionine-dependent methyltransferase activity; F:protein dimerization activity; F:caffeate O-methyltransferase activity | Caffeate O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| JG648265.1 | Tinospora cordifolia | basic form of pathogenesis-related protein 1-like | 482 | P:GO:0006952; P:GO:0009607; C:GO:0005576 | P:defense response; P:response to biotic stimulus; C:extracellular region | no GO terms | view details | |||
| JG646197.1 | Tinospora cordifolia | tumor-related protein | 557 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647312.1 | Tinospora cordifolia | auxin-induced protein AUX22 | 721 | P:GO:0006355; P:GO:0009734; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:auxin-activated signaling pathway; C:nucleus | P:regulation of transcription, DNA-templated; F:protein binding; C:nucleus | view details | |||
| JG647320.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase 3-like isoform X1 | 753 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG645542.1 | Tinospora cordifolia | pectinesterase 2-like | 771 | P:GO:0071555; F:GO:0030599; F:GO:0046910 | P:cell wall organization; F:pectinesterase activity; F:pectinesterase inhibitor activity | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG650940.1 | Tinospora cordifolia | adenosylhomocysteinase | 832 | P:GO:0006730; P:GO:0033353; F:GO:0004013; C:GO:0005829 | P:one-carbon metabolic process; P:S-adenosylmethionine cycle; F:adenosylhomocysteinase activity; C:cytosol | Adenosylhomocysteinase | no GO terms | view details | ||
| JG648701.1 | Tinospora cordifolia | tetrahydroberberine oxidase | 778 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649225.1 | Tinospora cordifolia | EH domain-containing protein 1 | 757 | P:GO:0006897; P:GO:0032456; P:GO:0042538; P:GO:0051260; F:GO:0005509; F:GO:0005525; C:GO:0005768; C:GO:0005886 | P:endocytosis; P:endocytic recycling; P:hyperosmotic salinity response; P:protein homooligomerization; F:calcium ion binding; F:GTP binding; C:endosome; C:plasma membrane | F:calcium ion binding; F:protein binding | view details | |||
| JG645659.1 | Tinospora cordifolia | peroxidase 12-like | 777 | P:GO:0006979; P:GO:0042744; P:GO:0098869; F:GO:0004601; F:GO:0020037; F:GO:0046872; C:GO:0005576; C:GO:0009505 | P:response to oxidative stress; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:peroxidase activity; F:heme binding; F:metal ion binding; C:extracellular region; C:plant-type cell wall | Acting on a peroxide as acceptor | P:response to oxidative stress; P:hydrogen peroxide catabolic process; F:peroxidase activity; F:heme binding | view details | ||
| JG648466.1 | Tinospora cordifolia | basic endochitinase-like | 821 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648904.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 671 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649695.1 | Tinospora cordifolia | probable prolyl 4-hydroxylase 9 | 811 | P:GO:0018401; F:GO:0004656; F:GO:0005506; F:GO:0031418; C:GO:0005789; C:GO:0016021 | P:peptidyl-proline hydroxylation to 4-hydroxy-L-proline; F:procollagen-proline 4-dioxygenase activity; F:iron ion binding; F:L-ascorbic acid binding; C:endoplasmic reticulum membrane; C:integral component of membrane | Procollagen-proline 4-dioxygenase | no GO terms | view details | ||
| JG646662.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 663 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649856.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 766 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648185.1 | Tinospora cordifolia | 40S ribosomal protein S4 | 730 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0022627 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic small ribosomal subunit | P:translation; F:RNA binding; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648022.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 766 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG646295.1 | Tinospora cordifolia | receptor-like serine/threonine-protein kinase SD1-8 | 488 | P:GO:0006468; P:GO:0048544; F:GO:0000166; F:GO:0004674; C:GO:0005886; C:GO:0016021 | P:protein phosphorylation; P:recognition of pollen; F:nucleotide binding; F:protein serine/threonine kinase activity; C:plasma membrane; C:integral component of membrane | Transferring phosphorus-containing groups | P:recognition of pollen | view details | ||
| JG650041.1 | Tinospora cordifolia | protein transport protein Sec24-like At3g07100 | 798 | P:GO:0006886; P:GO:0010584; P:GO:0048658; P:GO:0090110; F:GO:0000149; F:GO:0008270; C:GO:0030127; C:GO:0070971 | P:intracellular protein transport; P:pollen exine formation; P:anther wall tapetum development; P:COPII-coated vesicle cargo loading; F:SNARE binding; F:zinc ion binding; C:COPII vesicle coat; C:endoplasmic reticulum exit site | P:intracellular protein transport; P:endoplasmic reticulum to Golgi vesicle-mediated transport; F:zinc ion binding; C:COPII vesicle coat | view details | |||
| JG646388.1 | Tinospora cordifolia | CBL-interacting serine/threonine-protein kinase 14 | 826 | P:GO:0006468; P:GO:0007165; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310; C:GO:0016021 | P:protein phosphorylation; P:signal transduction; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity; C:integral component of membrane | Dual-specificity kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG650730.1 | Tinospora cordifolia | glyceraldehyde-3-phosphate dehydrogenase, cytosolic | 754 | P:GO:0006006; P:GO:0006096; F:GO:0004365; F:GO:0050661; F:GO:0051287; C:GO:0005829; C:GO:0009536 | P:glucose metabolic process; P:glycolytic process; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity; F:NADP binding; F:NAD binding; C:cytosol; C:plastid | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating); Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG647010.1 | Tinospora cordifolia | branched-chain-amino-acid aminotransferase 3, chloroplastic-like | 824 | P:GO:0008652; P:GO:0009082; F:GO:0050048; F:GO:0052654; F:GO:0052655; F:GO:0052656; C:GO:0016021 | P:cellular amino acid biosynthetic process; P:branched-chain amino acid biosynthetic process; F:L-leucine:2-oxoglutarate aminotransferase activity; F:L-leucine transaminase activity; F:L-valine transaminase activity; F:L-isoleucine transaminase activity; C:integral component of membrane | Leucine transaminase; Branched-chain-amino-acid transaminase | P:branched-chain amino acid metabolic process; F:catalytic activity; F:branched-chain-amino-acid transaminase activity | view details | ||
| JG647816.1 | Tinospora cordifolia | Polyphenol oxidase protein | 755 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:catechol oxidase activity; F:oxidoreductase activity | view details | ||
| JG649547.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 888 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| MG946872.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1413 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG646426.1 | Tinospora cordifolia | probable leucine-rich repeat receptor-like protein kinase At5g49770 | 807 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG646079.1 | Tinospora cordifolia | basic endochitinase | 796 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646688.1 | Tinospora cordifolia | peroxisomal fatty acid beta-oxidation multifunctional protein MFP2 | 749 | P:GO:0006635; F:GO:0003857; F:GO:0004165; F:GO:0004300; F:GO:0008692; F:GO:0070403; C:GO:0005777; C:GO:0016021 | P:fatty acid beta-oxidation; F:3-hydroxyacyl-CoA dehydrogenase activity; F:delta(3)-delta(2)-enoyl-CoA isomerase activity; F:enoyl-CoA hydratase activity; F:3-hydroxybutyryl-CoA epimerase activity; F:NAD+ binding; C:peroxisome; C:integral component of membrane | 3-hydroxyacyl-CoA dehydrogenase; Delta(3)-Delta(2)-enoyl-CoA isomerase; Enoyl-CoA hydratase; 3-hydroxybutyryl-CoA epimerase | P:fatty acid metabolic process; F:catalytic activity; F:NAD+ binding | view details | ||
| JG649219.1 | Tinospora cordifolia | auxin response factor 8 isoform X1 | 774 | P:GO:0006355; P:GO:0009734; P:GO:0009908; P:GO:0010154; F:GO:0000976; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:auxin-activated signaling pathway; P:flower development; P:fruit development; F:transcription cis-regulatory region binding; C:nucleus | P:regulation of transcription, DNA-templated; P:response to hormone; F:DNA binding; C:nucleus | view details | |||
| JG650728.1 | Tinospora cordifolia | cytochrome P450 714A1-like | 815 | F:GO:0004497; F:GO:0046872; C:GO:0016020 | F:monooxygenase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645961.1 | Tinospora cordifolia | Major intrinsic protein | 795 | P:GO:0006833; P:GO:0009414; P:GO:0055085; F:GO:0015250; C:GO:0005886; C:GO:0016021 | P:water transport; P:response to water deprivation; P:transmembrane transport; F:water channel activity; C:plasma membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:channel activity; C:membrane | view details | ||
| JG646846.1 | Tinospora cordifolia | glutathione S-transferase L3 | 528 | P:GO:0006749; P:GO:0010731; F:GO:0004364; C:GO:0016021 | P:glutathione metabolic process; P:protein glutathionylation; F:glutathione transferase activity; C:integral component of membrane | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG647321.1 | Tinospora cordifolia | aldehyde dehydrogenase family 2 member B7, mitochondrial-like | 792 | F:GO:0004029; F:GO:0043878 | F:aldehyde dehydrogenase (NAD+) activity; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity | Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG650382.1 | Tinospora cordifolia | cryptochrome DASH, chloroplastic/mitochondrial | 814 | P:GO:0000719; P:GO:0018298; F:GO:0003677; F:GO:0003913; F:GO:0071949 | P:photoreactive repair; P:obsolete protein-chromophore linkage; F:DNA binding; F:DNA photolyase activity; F:FAD binding | Carbon-carbon lyases | no GO terms | view details | ||
| MH569065.1 | Tinospora cordifolia | maturase K | 477 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG646162.1 | Tinospora cordifolia | probable glutathione S-transferase | 734 | P:GO:0006749; P:GO:0009636; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; P:response to toxic substance; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG647070.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 785 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649938.1 | Tinospora cordifolia | zinc finger CCCH domain-containing protein 53 isoform X1 | 735 | F:GO:0003677; F:GO:0003723; F:GO:0046872 | F:DNA binding; F:RNA binding; F:metal ion binding | F:nucleic acid binding; F:RNA binding; F:metal ion binding | view details | |||
| JG648163.1 | Tinospora cordifolia | CTP synthase | 834 | P:GO:0006541; P:GO:0019856; P:GO:0044210; F:GO:0003883; F:GO:0005524; F:GO:0042802; C:GO:0016021 | P:glutamine metabolic process; P:pyrimidine nucleobase biosynthetic process; P:de novo CTP biosynthetic process; F:CTP synthase activity; F:ATP binding; F:identical protein binding; C:integral component of membrane | CTP synthase (glutamine hydrolyzing) | P:pyrimidine nucleotide biosynthetic process; F:CTP synthase activity | view details | ||
| JG649991.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 764 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG648721.1 | Tinospora cordifolia | ankyrin repeat domain-containing protein, chloroplastic-like | 841 | P:GO:0090305; F:GO:0004518; C:GO:0009507 | P:nucleic acid phosphodiester bond hydrolysis; F:nuclease activity; C:chloroplast | F:protein binding | view details | |||
| JG650494.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 842 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG645635.1 | Tinospora cordifolia | probable esterase KAI2 | 852 | P:GO:0080167 | P:response to karrikin | no GO terms | view details | |||
| JG649850.1 | Tinospora cordifolia | U-box domain-containing protein 30-like | 741 | P:GO:0016567; P:GO:1900457; F:GO:0061630; C:GO:0005886 | P:protein ubiquitination; P:regulation of brassinosteroid mediated signaling pathway; F:ubiquitin protein ligase activity; C:plasma membrane | Transferases | P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:ubiquitin protein ligase activity | view details | ||
| JG650518.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 898 | P:GO:0005975; P:GO:0006952; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| MA258132.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 990 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG646343.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 808 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650788.1 | Tinospora cordifolia | monodehydroascorbate reductase | 811 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG645908.1 | Tinospora cordifolia | aspartate aminotransferase, cytoplasmic | 841 | P:GO:0006520; P:GO:0009058; F:GO:0004069; F:GO:0030170; C:GO:0005739 | P:cellular amino acid metabolic process; P:biosynthetic process; F:L-aspartate:2-oxoglutarate aminotransferase activity; F:pyridoxal phosphate binding; C:mitochondrion | Aspartate transaminase | P:cellular amino acid metabolic process; P:biosynthetic process; F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG647686.1 | Tinospora cordifolia | Cysteine-rich receptor-like protein kinase 10 | 852 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG647348.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 734 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647199.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 806 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG647689.1 | Tinospora cordifolia | heat shock 70 kDa protein 15-like | 773 | P:GO:0006457; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0005829 | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:cytosol | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG645954.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 827 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650943.1 | Tinospora cordifolia | calmodulin-binding protein 60 B-like | 724 | P:GO:0006355; P:GO:0080142; F:GO:0003700; F:GO:0005516; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:regulation of salicylic acid biosynthetic process; F:DNA-binding transcription factor activity; F:calmodulin binding; F:sequence-specific DNA binding; C:nucleus | F:calmodulin binding | view details | |||
| JG646214.1 | Tinospora cordifolia | FAD linked oxidase | 416 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650877.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 857 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649561.1 | Tinospora cordifolia | acetyl-CoA carboxylase 1-like | 834 | P:GO:0006633; P:GO:2001295; F:GO:0003989; F:GO:0005524; F:GO:0016740; F:GO:0046872; C:GO:0009536 | P:fatty acid biosynthetic process; P:malonyl-CoA biosynthetic process; F:acetyl-CoA carboxylase activity; F:ATP binding; F:transferase activity; F:metal ion binding; C:plastid | Transferases; Acetyl-CoA carboxylase | no GO terms | view details | ||
| JG650423.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 802 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649831.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase 1-like | 745 | P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG648353.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 735 | P:GO:0005975; F:GO:0016765; F:GO:0042973 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647094.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 784 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| LY749003.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 990 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG646096.1 | Tinospora cordifolia | laccase-7-like | 884 | P:GO:0046274; F:GO:0005507; F:GO:0052716; C:GO:0048046 | P:lignin catabolic process; F:copper ion binding; F:hydroquinone:oxygen oxidoreductase activity; C:apoplast | Laccase | F:copper ion binding | view details | ||
| JG645555.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 794 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650841.1 | Tinospora cordifolia | probable galacturonosyltransferase 9 | 791 | P:GO:0045489; P:GO:0071555; F:GO:0047262; C:GO:0000139; C:GO:0016021 | P:pectin biosynthetic process; P:cell wall organization; F:polygalacturonate 4-alpha-galacturonosyltransferase activity; C:Golgi membrane; C:integral component of membrane | Polygalacturonate 4-alpha-galacturonosyltransferase | F:polygalacturonate 4-alpha-galacturonosyltransferase activity | view details | ||
| JG650602.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 848 | F:GO:0050660 | F:flavin adenine dinucleotide binding | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | |||
| JG646239.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 760 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649797.1 | Tinospora cordifolia | auxin response factor 8-like isoform X2 | 703 | P:GO:0006355; P:GO:0009734; P:GO:0048608; F:GO:0003677; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:auxin-activated signaling pathway; P:reproductive structure development; F:DNA binding; C:nucleus | P:regulation of transcription, DNA-templated; P:response to hormone; F:DNA binding | view details | |||
| JG649539.1 | Tinospora cordifolia | protein EXORDIUM-like 2 | 769 | P:GO:0090502; F:GO:0003676; F:GO:0004523; C:GO:0005615; C:GO:0016021 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:nucleic acid binding; F:RNA-DNA hybrid ribonuclease activity; C:extracellular space; C:integral component of membrane | Acting on ester bonds; Ribonuclease H | no GO terms | view details | ||
| JG646281.1 | Tinospora cordifolia | Glucan endo-1,3-beta-glucosidase | 549 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645614.1 | Tinospora cordifolia | heat shock 70 kDa protein, mitochondrial | 805 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005739 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:mitochondrion | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG650739.1 | Tinospora cordifolia | ATP-binding cassette domain-containing protein | 801 | P:GO:0055085; F:GO:0005524; F:GO:0140359; C:GO:0016021 | P:transmembrane transport; F:ATP binding; F:ABC-type transporter activity; C:integral component of membrane | Translocases | F:ATP binding | view details | ||
| JG647894.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 779 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650033.1 | Tinospora cordifolia | malate dehydrogenase | 740 | P:GO:0006099; P:GO:0006107; P:GO:0006108; P:GO:0006734; F:GO:0030060 | P:tricarboxylic acid cycle; P:oxaloacetate metabolic process; P:malate metabolic process; P:NADH metabolic process; F:L-malate dehydrogenase activity | Malate dehydrogenase | P:malate metabolic process; F:catalytic activity; F:oxidoreductase activity; F:malate dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG648971.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 559 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650026.1 | Tinospora cordifolia | 6-phosphogluconate dehydrogenase | 800 | P:GO:0009051; P:GO:0046177; F:GO:0004616; F:GO:0050661; C:GO:0005829 | P:pentose-phosphate shunt, oxidative branch; P:D-gluconate catabolic process; F:phosphogluconate dehydrogenase (decarboxylating) activity; F:NADP binding; C:cytosol | Phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | P:pentose-phosphate shunt; F:phosphogluconate dehydrogenase (decarboxylating) activity; F:oxidoreductase activity; F:NADP binding | view details | ||
| JG646461.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 769 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648876.1 | Tinospora cordifolia | caffeic acid 3-O-methyltransferase-like | 653 | P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757; F:GO:0046983 | P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| MT850046.1 | Tinospora cordifolia | ATP synthase CF1 beta subunit | 1497 | P:GO:0015986; P:GO:1902600; F:GO:0005524; F:GO:0046933; F:GO:0046961; C:GO:0009535; C:GO:0045261 | P:proton motive force-driven ATP synthesis; P:proton transmembrane transport; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:chloroplast thylakoid membrane; C:proton-transporting ATP synthase complex, catalytic core F(1) | H(+)-exporting diphosphatase; H(+)-transporting two-sector ATPase; Ligases | P:proton motive force-driven ATP synthesis; P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:proton-transporting ATP synthase complex, catalytic core F(1) | view details | ||
| JG648031.1 | Tinospora cordifolia | glyceraldehyde-3-phosphate dehydrogenase, cytosolic | 845 | P:GO:0006006; P:GO:0006096; F:GO:0004365; F:GO:0050661; F:GO:0051287; C:GO:0005829; C:GO:0009536 | P:glucose metabolic process; P:glycolytic process; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity; F:NADP binding; F:NAD binding; C:cytosol; C:plastid | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating); Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG646274.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 659 | F:GO:0016491; F:GO:0071949; C:GO:0005576 | F:oxidoreductase activity; F:FAD binding; C:extracellular region | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647114.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 781 | P:GO:0005975; P:GO:0006952; F:GO:0004553; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:anchored component of plasma membrane | Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647590.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 818 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG650685.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1-like | 807 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | no GO terms | view details | ||
| JG650117.1 | Tinospora cordifolia | 6-phosphogluconate dehydrogenase | 798 | P:GO:0009051; P:GO:0046177; F:GO:0004616; F:GO:0050661; C:GO:0005829 | P:pentose-phosphate shunt, oxidative branch; P:D-gluconate catabolic process; F:phosphogluconate dehydrogenase (decarboxylating) activity; F:NADP binding; C:cytosol | Phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | P:pentose-phosphate shunt; F:phosphogluconate dehydrogenase (decarboxylating) activity; F:oxidoreductase activity; F:NADP binding | view details | ||
| JG650525.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 861 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649066.1 | Tinospora cordifolia | beta-amylase 8 | 829 | P:GO:0000272; P:GO:0010468; F:GO:0016161; F:GO:0102229 | P:polysaccharide catabolic process; P:regulation of gene expression; F:beta-amylase activity; F:amylopectin maltohydrolase activity | Beta-amylase | P:polysaccharide catabolic process; F:beta-amylase activity | view details | ||
| JG647528.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 712 | P:GO:0005975; F:GO:0004497; F:GO:0004553; F:GO:0016705; F:GO:0016765; F:GO:0046872; C:GO:0046658 | P:carbohydrate metabolic process; F:monooxygenase activity; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:metal ion binding; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647937.1 | Tinospora cordifolia | Xanthotoxin 5-hydroxylase CYP82C4 | 706 | P:GO:0019438; P:GO:0098542; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:defense response to other organism; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646983.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 130 | P:GO:0009086; P:GO:0009908; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:flower development; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | no IPS match | view details | ||
| JG648032.1 | Tinospora cordifolia | L-ascorbate peroxidase, cytosolic | 809 | P:GO:0009723; P:GO:0034599; P:GO:0042542; P:GO:0042744; P:GO:0045454; P:GO:0090378; P:GO:0098869; F:GO:0016688; F:GO:0020037; F:GO:0046872; C:GO:0009507 | P:response to ethylene; P:cellular response to oxidative stress; P:response to hydrogen peroxide; P:hydrogen peroxide catabolic process; P:cell redox homeostasis; P:seed trichome elongation; P:cellular oxidant detoxification; F:L-ascorbate peroxidase activity; F:heme binding; F:metal ion binding; C:chloroplast | L-ascorbate peroxidase | P:response to oxidative stress; P:cellular response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG649266.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 749 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650216.1 | Tinospora cordifolia | T-complex protein 1 subunit gamma | 712 | P:GO:0006457; F:GO:0005524; F:GO:0016887; F:GO:0051082; C:GO:0005832 | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; F:unfolded protein binding; C:chaperonin-containing T-complex | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG648386.1 | Tinospora cordifolia | Beta-glucosidase 18 | 833 | P:GO:0008152; F:GO:0008422; C:GO:0016020 | P:metabolic process; F:beta-glucosidase activity; C:membrane | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647243.1 | Tinospora cordifolia | acyltransferase-like protein At1g54570, chloroplastic | 836 | P:GO:0009820; F:GO:0004144 | P:alkaloid metabolic process; F:diacylglycerol O-acyltransferase activity | Diacylglycerol O-acyltransferase | no GO terms | view details | ||
| JG646114.1 | Tinospora cordifolia | dnaJ protein homolog | 804 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG649150.1 | Tinospora cordifolia | polygalacturonase-inhibiting protein | 795 | C:GO:0110165 | C:cellular anatomical entity | F:protein binding | view details | |||
| MS688301.1 | Tinospora cordifolia | Papaver somniferum O-methyltransferase 1 | 1074 | P:GO:0009708; P:GO:0032259; F:GO:0008171; F:GO:0030777; F:GO:0046983 | P:benzyl isoquinoline alkaloid biosynthetic process; P:methylation; F:O-methyltransferase activity; F:(S)-scoulerine 9-O-methyltransferase activity; F:protein dimerization activity | (S)-scoulerine 9-O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG647899.1 | Tinospora cordifolia | Linoleate 9S-lipoxygenase 5 | 783 | P:GO:0006633; P:GO:0031408; P:GO:0034440; F:GO:0016702; F:GO:0046872; C:GO:0005737 | P:fatty acid biosynthetic process; P:oxylipin biosynthetic process; P:lipid oxidation; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; F:metal ion binding; C:cytoplasm | Acting on single donors with incorporation of molecular oxygen (oxygenases). The oxygen incorporated need not be derived from O(2) | F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; F:metal ion binding | view details | ||
| JG649364.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 748 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG646382.1 | Tinospora cordifolia | 60S ribosomal protein L7-2-like | 794 | P:GO:0000463; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG648347.1 | Tinospora cordifolia | probable galacturonosyltransferase 9 | 864 | P:GO:0045489; P:GO:0071555; F:GO:0047262; C:GO:0000139; C:GO:0016021 | P:pectin biosynthetic process; P:cell wall organization; F:polygalacturonate 4-alpha-galacturonosyltransferase activity; C:Golgi membrane; C:integral component of membrane | Polygalacturonate 4-alpha-galacturonosyltransferase | F:polygalacturonate 4-alpha-galacturonosyltransferase activity | view details | ||
| JG648940.1 | Tinospora cordifolia | basic endochitinase-like | 748 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648979.1 | Tinospora cordifolia | Secretory carrier-associated membrane protein 1 | 214 | P:GO:0015031; C:GO:0005886; C:GO:0016021; C:GO:0030658; C:GO:0032588; C:GO:0055038 | P:protein transport; C:plasma membrane; C:integral component of membrane; C:transport vesicle membrane; C:trans-Golgi network membrane; C:recycling endosome membrane | P:protein transport; C:integral component of membrane | view details | |||
| JG649186.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At5g25630 | 754 | F:protein binding | view details | |||||
| JG648589.1 | Tinospora cordifolia | basic endochitinase-like | 850 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649970.1 | Tinospora cordifolia | pathogen-related protein-like | 220 | no GO terms | view details | |||||
| JG650793.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 679 | P:GO:0009058; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650771.1 | Tinospora cordifolia | Cytochrome P450 82A3 | 780 | P:GO:0098542; F:GO:0005488; F:GO:0016491 | P:defense response to other organism; F:binding; F:oxidoreductase activity | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648480.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 780 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650586.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 744 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650413.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 788 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649077.1 | Tinospora cordifolia | auxin response factor 2A-like | 776 | P:GO:0006355; P:GO:0009725; P:GO:0009734; F:GO:0003677; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:response to hormone; P:auxin-activated signaling pathway; F:DNA binding; C:nucleus | P:regulation of transcription, DNA-templated; P:response to hormone; F:DNA binding; F:protein binding | view details | |||
| JG645648.1 | Tinospora cordifolia | ---NA--- | 844 | P:proteolysis; F:aspartic-type endopeptidase activity | view details | |||||
| JG648258.1 | Tinospora cordifolia | Corytuberine synthase | 496 | P:GO:0009820; P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0005783; C:GO:0016020 | P:alkaloid metabolic process; P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:endoplasmic reticulum; C:membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647285.1 | Tinospora cordifolia | 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 | 529 | P:GO:0008299; P:GO:0016126; F:GO:0004420; C:GO:0005778; C:GO:0005789 | P:isoprenoid biosynthetic process; P:sterol biosynthetic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; C:peroxisomal membrane; C:endoplasmic reticulum membrane | Hydroxymethylglutaryl-CoA reductase (NADPH) | P:coenzyme A metabolic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity | view details | ||
| MK244347.1 | Tinospora cordifolia | maturase K | 892 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG648636.1 | Tinospora cordifolia | berberine bridge enzyme-like 26 | 785 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647760.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein At5g02240-like | 725 | F:GO:0016491 | F:oxidoreductase activity | Oxidoreductases | F:oxidoreductase activity | view details | ||
| MS688353.1 | Tinospora cordifolia | salutaridine reductase-like | 915 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG649423.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 766 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647384.1 | Tinospora cordifolia | ABC transporter I family member 20 | 676 | F:GO:0005524 | F:ATP binding | F:ATP binding | view details | |||
| JG647493.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 723 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650323.1 | Tinospora cordifolia | polyphenol oxidase | 812 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG645933.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 668 | P:GO:0005975; P:GO:0006952; F:GO:0005506; F:GO:0016712; F:GO:0016765; F:GO:0020037; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:heme binding; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648172.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 783 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646536.1 | Tinospora cordifolia | beta-glucosidase BoGH3B-like | 752 | P:GO:0009251; F:GO:0008422; C:GO:0005576 | P:glucan catabolic process; F:beta-glucosidase activity; C:extracellular region | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648797.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 764 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648399.1 | Tinospora cordifolia | basic endochitinase-like | 822 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648243.1 | Tinospora cordifolia | methylecgonone reductase-like | 423 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG646146.1 | Tinospora cordifolia | UTP--glucose-1-phosphate uridylyltransferase | 858 | P:GO:0005977; P:GO:0006011; F:GO:0003983; C:GO:0005737; C:GO:0016021 | P:glycogen metabolic process; P:UDP-glucose metabolic process; F:UTP:glucose-1-phosphate uridylyltransferase activity; C:cytoplasm; C:integral component of membrane | UTP-monosaccharide-1-phosphate uridylyltransferase; UTP--glucose-1-phosphate uridylyltransferase | P:UDP-glucose metabolic process; F:UTP:glucose-1-phosphate uridylyltransferase activity; F:uridylyltransferase activity | view details | ||
| JG650503.1 | Tinospora cordifolia | aspartate--tRNA ligase 2, cytoplasmic-like | 822 | P:GO:0006422; P:GO:0016104; F:GO:0003677; F:GO:0003723; F:GO:0004815; F:GO:0005524; F:GO:0016866; C:GO:0005811; C:GO:0005829; C:GO:0017101 | P:aspartyl-tRNA aminoacylation; P:triterpenoid biosynthetic process; F:DNA binding; F:RNA binding; F:aspartate-tRNA ligase activity; F:ATP binding; F:intramolecular transferase activity; C:lipid droplet; C:cytosol; C:aminoacyl-tRNA synthetase multienzyme complex | Intramolecular transferases; Aspartate--tRNA ligase | P:aspartyl-tRNA aminoacylation; F:nucleotide binding; F:nucleic acid binding; F:aspartate-tRNA ligase activity; F:ATP binding; C:cytoplasm | view details | ||
| JG647906.1 | Tinospora cordifolia | transcription factor GTE10-like isoform X1 | 738 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG648182.1 | Tinospora cordifolia | ras-related protein RABA6b-like | 669 | P:GO:0006886; F:GO:0003924; F:GO:0005525; C:GO:0005768 | P:intracellular protein transport; F:GTPase activity; F:GTP binding; C:endosome | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG649179.1 | Tinospora cordifolia | polygalacturonase inhibitor | 561 | C:GO:0016020 | C:membrane | F:protein binding | view details | |||
| JG649896.1 | Tinospora cordifolia | catalase-2 isoform X2 | 437 | P:GO:0042542; P:GO:0042744; P:GO:0098869; F:GO:0004096; F:GO:0020037; F:GO:0046872; C:GO:0005777; C:GO:0005886 | P:response to hydrogen peroxide; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:catalase activity; F:heme binding; F:metal ion binding; C:peroxisome; C:plasma membrane | Catalase | P:response to oxidative stress; F:catalase activity; F:heme binding | view details | ||
| JG649353.1 | Tinospora cordifolia | beta-glucosidase 12-like | 641 | P:GO:0005975; P:GO:0009651; P:GO:0019762; F:GO:0008422 | P:carbohydrate metabolic process; P:response to salt stress; P:glucosinolate catabolic process; F:beta-glucosidase activity | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649470.1 | Tinospora cordifolia | aquaporin PIP2-7 | 574 | P:GO:0006833; P:GO:0055085; F:GO:0015250; C:GO:0005886; C:GO:0016021 | P:water transport; P:transmembrane transport; F:water channel activity; C:plasma membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:channel activity; C:membrane | view details | ||
| JG650681.1 | Tinospora cordifolia | putative clathrin assembly protein At2g01600 isoform X1 | 827 | P:GO:0048268; P:GO:2000369; F:GO:0005545; F:GO:0030276; C:GO:0005794; C:GO:0005905; C:GO:0030136 | P:clathrin coat assembly; P:regulation of clathrin-dependent endocytosis; F:1-phosphatidylinositol binding; F:clathrin binding; C:Golgi apparatus; C:clathrin-coated pit; C:clathrin-coated vesicle | P:clathrin coat assembly; P:clathrin-dependent endocytosis; F:phospholipid binding; F:1-phosphatidylinositol binding; F:clathrin binding; C:clathrin-coated vesicle | view details | |||
| JG645715.1 | Tinospora cordifolia | arginine decarboxylase-like | 670 | P:GO:0006527; P:GO:0008295; P:GO:0033388; F:GO:0008792 | P:arginine catabolic process; P:spermidine biosynthetic process; P:putrescine biosynthetic process from arginine; F:arginine decarboxylase activity | Arginine decarboxylase | P:arginine catabolic process; P:spermidine biosynthetic process; F:catalytic activity; F:arginine decarboxylase activity | view details | ||
| JG649360.1 | Tinospora cordifolia | corytuberine synthase | 823 | P:GO:0009820; P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016717; F:GO:0020037; C:GO:0005783; C:GO:0016020 | P:alkaloid metabolic process; P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water; F:heme binding; C:endoplasmic reticulum; C:membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647989.1 | Tinospora cordifolia | 40S ribosomal protein S4 | 736 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0022627 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic small ribosomal subunit | P:translation; F:RNA binding; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649346.1 | Tinospora cordifolia | putative methylesterase 11, chloroplastic | 755 | P:GO:0009694; P:GO:0009696; F:GO:0080030; F:GO:0080031; F:GO:0080032 | P:jasmonic acid metabolic process; P:salicylic acid metabolic process; F:methyl indole-3-acetate esterase activity; F:methyl salicylate esterase activity; F:methyl jasmonate esterase activity | Carboxylesterase | no GO terms | view details | ||
| JG648058.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 850 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650948.1 | Tinospora cordifolia | proteasome subunit alpha type-5 | 781 | P:GO:0043161; C:GO:0005634; C:GO:0005737; C:GO:0019773 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; C:nucleus; C:cytoplasm; C:proteasome core complex, alpha-subunit complex | P:ubiquitin-dependent protein catabolic process; P:proteasome-mediated ubiquitin-dependent protein catabolic process; P:proteolysis involved in protein catabolic process; C:proteasome core complex; C:proteasome core complex, alpha-subunit complex | view details | |||
| JG649537.1 | Tinospora cordifolia | Tubulin beta-1 chain | 794 | P:GO:0000226; P:GO:0000278; F:GO:0003729; F:GO:0003924; F:GO:0005200; F:GO:0005515; F:GO:0005525; C:GO:0005737; C:GO:0005874 | P:microtubule cytoskeleton organization; P:mitotic cell cycle; F:mRNA binding; F:GTPase activity; F:structural constituent of cytoskeleton; F:protein binding; F:GTP binding; C:cytoplasm; C:microtubule | Acting on acid anhydrides | P:microtubule-based process; F:structural constituent of cytoskeleton; F:GTP binding; C:microtubule | view details | ||
| JG649530.1 | Tinospora cordifolia | ABC transporter G family member 39 | 706 | P:GO:0055085; F:GO:0005524; F:GO:0140359; C:GO:0005886; C:GO:0016021 | P:transmembrane transport; F:ATP binding; F:ABC-type transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | C:membrane | view details | ||
| JG650467.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 777 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647806.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 836 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650190.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 808 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650246.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase-like | 783 | F:GO:0016491; F:GO:0046872 | F:oxidoreductase activity; F:metal ion binding | Oxidoreductases | no GO terms | view details | ||
| JG648056.1 | Tinospora cordifolia | protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 | 873 | P:GO:0006338; F:GO:0003824; F:GO:0005488; F:GO:0140657 | P:chromatin remodeling; F:catalytic activity; F:binding; F:ATP-dependent activity | no IPS match | view details | |||
| JG648949.1 | Tinospora cordifolia | probable WRKY transcription factor 47 | 822 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG646520.1 | Tinospora cordifolia | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like | 832 | P:GO:0006099; P:GO:0033512; F:GO:0004149; C:GO:0045252 | P:tricarboxylic acid cycle; P:L-lysine catabolic process to acetyl-CoA via saccharopine; F:dihydrolipoyllysine-residue succinyltransferase activity; C:oxoglutarate dehydrogenase complex | Dihydrolipoyllysine-residue succinyltransferase | no GO terms | view details | ||
| JG650201.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 785 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG648899.1 | Tinospora cordifolia | probable inactive purple acid phosphatase 2 | 647 | P:GO:0016311; F:GO:0003993; F:GO:0046872; C:GO:0016021 | P:dephosphorylation; F:acid phosphatase activity; F:metal ion binding; C:integral component of membrane | Acid phosphatase | no GO terms | view details | ||
| JG645963.1 | Tinospora cordifolia | probable CoA ligase CCL7 | 746 | P:GO:0009698; F:GO:0016207; C:GO:0005737; C:GO:0016021 | P:phenylpropanoid metabolic process; F:4-coumarate-CoA ligase activity; C:cytoplasm; C:integral component of membrane | 4-coumarate--CoA ligase | no GO terms | view details | ||
| JG646707.1 | Tinospora cordifolia | polyubiquitin isoform X1 | 808 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG650028.1 | Tinospora cordifolia | basic endochitinase-like | 724 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647306.1 | Tinospora cordifolia | pyrophosphate-energized vacuolar membrane proton pump | 779 | P:GO:1902600; F:GO:0004427; F:GO:0009678; C:GO:0016021 | P:proton transmembrane transport; F:inorganic diphosphatase activity; F:pyrophosphate hydrolysis-driven proton transmembrane transporter activity; C:integral component of membrane | H(+)-exporting diphosphatase; Inorganic diphosphatase | P:proton transmembrane transport; F:inorganic diphosphatase activity; F:pyrophosphate hydrolysis-driven proton transmembrane transporter activity; C:membrane | view details | ||
| JG650950.1 | Tinospora cordifolia | basic endochitinase | 794 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646643.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 788 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649908.1 | Tinospora cordifolia | cytochrome P450 81Q32-like | 323 | P:GO:0098542; F:GO:0016709; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen; F:metal ion binding; C:membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| LY340343.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 966 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG650686.1 | Tinospora cordifolia | protein FAR1-RELATED SEQUENCE 3 | 800 | P:GO:0006355; F:GO:0008270; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:zinc ion binding; C:nucleus | P:regulation of transcription, DNA-templated; F:zinc ion binding | view details | |||
| JG645923.1 | Tinospora cordifolia | cytochrome P450 81Q32-like | 847 | P:GO:0009058; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| MH538119.1 | Tinospora cordifolia | Reticuline oxidase | 979 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | ||
| JG650887.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 783 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647842.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 752 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG645527.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 821 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG645706.1 | Tinospora cordifolia | malate dehydrogenase | 755 | P:GO:0006099; P:GO:0006107; P:GO:0006108; P:GO:0006734; F:GO:0030060 | P:tricarboxylic acid cycle; P:oxaloacetate metabolic process; P:malate metabolic process; P:NADH metabolic process; F:L-malate dehydrogenase activity | Malate dehydrogenase | P:malate metabolic process; F:catalytic activity; F:oxidoreductase activity; F:malate dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG645641.1 | Tinospora cordifolia | monodehydroascorbate reductase | 805 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG645608.1 | Tinospora cordifolia | vacuolar H+-ATPase | 643 | P:GO:0015986; P:GO:1902600; F:GO:0005524; F:GO:0016787; F:GO:0046933; F:GO:0046961; C:GO:0031090; C:GO:0033180 | P:proton motive force-driven ATP synthesis; P:proton transmembrane transport; F:ATP binding; F:hydrolase activity; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:organelle membrane; C:proton-transporting V-type ATPase, V1 domain | H(+)-exporting diphosphatase; H(+)-transporting two-sector ATPase; Hydrolases; Ligases | P:ATP metabolic process; F:ATP binding; F:proton-transporting ATPase activity, rotational mechanism | view details | ||
| JG647504.1 | Tinospora cordifolia | Aconitate hydratase | 738 | P:GO:0006099; P:GO:0006101; F:GO:0003994; F:GO:0046872; F:GO:0047780; F:GO:0051539; C:GO:0005739; C:GO:0005829 | P:tricarboxylic acid cycle; P:citrate metabolic process; F:aconitate hydratase activity; F:metal ion binding; F:citrate dehydratase activity; F:4 iron, 4 sulfur cluster binding; C:mitochondrion; C:cytosol | Aconitate hydratase | no GO terms | view details | ||
| JG649296.1 | Tinospora cordifolia | copper-transporting ATPase HMA4-like | 807 | P:GO:0035434; P:GO:0055070; F:GO:0005507; F:GO:0005524; F:GO:0016887; F:GO:0043682; C:GO:0016021 | P:copper ion transmembrane transport; P:copper ion homeostasis; F:copper ion binding; F:ATP binding; F:ATP hydrolysis activity; F:P-type divalent copper transporter activity; C:integral component of membrane | Acting on acid anhydrides; P-type Cu(2+) transporter | F:nucleotide binding; F:transporter activity; F:ATP binding; F:ATP hydrolysis activity; C:integral component of membrane | view details | ||
| JG647669.1 | Tinospora cordifolia | NADH-cytochrome b5 reductase-like protein | 851 | F:GO:0004128; C:GO:0016021 | F:cytochrome-b5 reductase activity, acting on NAD(P)H; C:integral component of membrane | Cytochrome-b5 reductase | F:oxidoreductase activity | view details | ||
| JG649931.1 | Tinospora cordifolia | histone H3 | 320 | F:GO:0003677; F:GO:0046982; C:GO:0000786; C:GO:0005634 | F:DNA binding; F:protein heterodimerization activity; C:nucleosome; C:nucleus | no IPS match | view details | |||
| JG649002.1 | Tinospora cordifolia | autophagy-related protein 8c | 210 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016020 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:membrane | no GO terms | view details | |||
| JG649611.1 | Tinospora cordifolia | pathogen-related protein-like | 518 | no GO terms | view details | |||||
| JG649483.1 | Tinospora cordifolia | berberine bridge enzyme-like 18 | 832 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646532.1 | Tinospora cordifolia | serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | 809 | P:GO:0006470; P:GO:0050790; F:GO:0019888; C:GO:0000159; C:GO:0005737 | P:protein dephosphorylation; P:regulation of catalytic activity; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex; C:cytoplasm | no GO terms | view details | |||
| JG648241.1 | Tinospora cordifolia | 40S ribosomal protein S7 | 570 | P:GO:0006364; P:GO:0006412; P:GO:0042274; F:GO:0003735; C:GO:0022627; C:GO:0032040 | P:rRNA processing; P:translation; P:ribosomal small subunit biogenesis; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit; C:small-subunit processome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647477.1 | Tinospora cordifolia | Alpha-dioxygenase 1 | 838 | P:GO:0006979; P:GO:0098869; F:GO:0004601; F:GO:0020037; F:GO:0051213 | P:response to oxidative stress; P:cellular oxidant detoxification; F:peroxidase activity; F:heme binding; F:dioxygenase activity | Acting on a peroxide as acceptor | P:response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG646438.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 838 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649935.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 406 | F:GO:0004097; F:GO:0046872 | F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | no GO terms | view details | ||
| JG647464.1 | Tinospora cordifolia | thaumatin-like protein | 127 | P:GO:0006952 | P:defense response | no GO terms | view details | |||
| JG647014.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 823 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG645677.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 766 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| MS688300.1 | Tinospora cordifolia | (RS)-norcoclaurine 6-O-methyltransferase-like | 1050 | P:GO:0008152; F:GO:0008757 | P:metabolic process; F:S-adenosylmethionine-dependent methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG650922.1 | Tinospora cordifolia | zinc finger CCCH domain-containing protein 53 isoform X1 | 797 | F:GO:0003677; F:GO:0003723; F:GO:0046872 | F:DNA binding; F:RNA binding; F:metal ion binding | F:nucleic acid binding; F:RNA binding; F:metal ion binding | view details | |||
| JG649644.1 | Tinospora cordifolia | cationic amino acid transporter 1-like | 767 | P:GO:0015813; P:GO:1903401; F:GO:0005313; F:GO:0015189; C:GO:0005886; C:GO:0016021 | P:L-glutamate transmembrane transport; P:L-lysine transmembrane transport; F:L-glutamate transmembrane transporter activity; F:L-lysine transmembrane transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| JG645600.1 | Tinospora cordifolia | basic endochitinase-like | 643 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061; C:GO:0005576; C:GO:0005773 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding; C:extracellular region; C:vacuole | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650051.1 | Tinospora cordifolia | methionine S-methyltransferase | 762 | P:GO:0001887; P:GO:0032259; P:GO:0046500; P:GO:1901566; F:GO:0008483; F:GO:0030170; F:GO:0030732 | P:selenium compound metabolic process; P:methylation; P:S-adenosylmethionine metabolic process; P:organonitrogen compound biosynthetic process; F:transaminase activity; F:pyridoxal phosphate binding; F:methionine S-methyltransferase activity | Transferring nitrogenous groups; Methionine S-methyltransferase | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG646904.1 | Tinospora cordifolia | linamarin synthase 2-like | 654 | P:GO:0009718; F:GO:0047213; F:GO:0080043; F:GO:0080044 | P:anthocyanin-containing compound biosynthetic process; F:anthocyanidin 3-O-glucosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Anthocyanidin 3-O-glucosyltransferase | no GO terms | view details | ||
| JG650433.1 | Tinospora cordifolia | hypersensitive-induced reaction 1 protein | 691 | C:GO:0005737; C:GO:0043231 | C:cytoplasm; C:intracellular membrane-bounded organelle | no GO terms | view details | |||
| JG647824.1 | Tinospora cordifolia | methyl-CpG-binding domain-containing protein 9-like | 726 | P:GO:0006325; P:GO:0016570; F:GO:0003677; F:GO:0046872; C:GO:0005634 | P:chromatin organization; P:histone modification; F:DNA binding; F:metal ion binding; C:nucleus | no GO terms | view details | |||
| JG645843.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 795 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648995.1 | Tinospora cordifolia | guanosine nucleotide diphosphate dissociation inhibitor 2 | 560 | P:GO:0007264; P:GO:0015031; P:GO:0016192; P:GO:0050790; F:GO:0005093; C:GO:0005737 | P:small GTPase mediated signal transduction; P:protein transport; P:vesicle-mediated transport; P:regulation of catalytic activity; F:Rab GDP-dissociation inhibitor activity; C:cytoplasm | P:small GTPase mediated signal transduction; P:protein transport; F:GDP-dissociation inhibitor activity; F:Rab GDP-dissociation inhibitor activity | view details | |||
| JG650559.1 | Tinospora cordifolia | guanosine nucleotide diphosphate dissociation inhibitor 2 | 835 | P:GO:0007264; P:GO:0015031; P:GO:0016192; P:GO:0050790; F:GO:0005093; C:GO:0005737 | P:small GTPase mediated signal transduction; P:protein transport; P:vesicle-mediated transport; P:regulation of catalytic activity; F:Rab GDP-dissociation inhibitor activity; C:cytoplasm | P:small GTPase mediated signal transduction; P:protein transport; F:GDP-dissociation inhibitor activity; F:Rab GDP-dissociation inhibitor activity | view details | |||
| JG649816.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 747 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG648938.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 735 | F:GO:0016491; F:GO:0071949; C:GO:0005576 | F:oxidoreductase activity; F:FAD binding; C:extracellular region | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647655.1 | Tinospora cordifolia | FAD linked oxidase | 777 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| MS687900.1 | Tinospora cordifolia | protein SRG1-like | 903 | P:GO:1901576; F:GO:0016705; F:GO:0046872 | P:organic substance biosynthetic process; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:metal ion binding | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no GO terms | view details | ||
| JG647409.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 777 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647036.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 846 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG646138.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 684 | P:GO:0009058; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647625.1 | Tinospora cordifolia | GDSL esterase/lipase 7-like | 836 | F:GO:0016788; C:GO:0016020 | F:hydrolase activity, acting on ester bonds; C:membrane | Acting on ester bonds | F:hydrolase activity, acting on ester bonds | view details | ||
| JG648376.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 863 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG648429.1 | Tinospora cordifolia | Golgi to ER traffic protein 4 homolog | 810 | P:GO:0045048; P:GO:0048767; C:GO:0005829 | P:protein insertion into ER membrane; P:root hair elongation; C:cytosol | P:protein insertion into ER membrane; F:protein binding | view details | |||
| JG650225.1 | Tinospora cordifolia | 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like | 753 | F:GO:0047568 | F:3-oxo-5-beta-steroid 4-dehydrogenase activity | 3-oxo-5-beta-steroid 4-dehydrogenase | no GO terms | view details | ||
| JG645898.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like | 747 | F:GO:0016491; F:GO:0046872 | F:oxidoreductase activity; F:metal ion binding | Oxidoreductases | no GO terms | view details | ||
| JG650590.1 | Tinospora cordifolia | Plastid-lipid-associated protein, chloroplastic | 802 | F:GO:0016491; C:GO:0009536 | F:oxidoreductase activity; C:plastid | Oxidoreductases | no GO terms | view details | ||
| JG647915.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase 14 | 704 | P:GO:0005975; F:GO:0042973; C:GO:0016021; C:GO:0046658 | P:carbohydrate metabolic process; F:glucan endo-1,3-beta-D-glucosidase activity; C:integral component of membrane; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646722.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 847 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648184.1 | Tinospora cordifolia | 40S ribosomal protein S4 | 227 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0022627 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic small ribosomal subunit | P:translation; F:RNA binding; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649629.1 | Tinospora cordifolia | ACT domain-containing protein ACR6-like | 825 | no GO terms | view details | |||||
| JG650932.1 | Tinospora cordifolia | phosphatidylinositol 4-kinase gamma 4-like | 769 | P:GO:0016310; P:GO:0046854; F:GO:0004430 | P:phosphorylation; P:phosphatidylinositol phosphate biosynthetic process; F:1-phosphatidylinositol 4-kinase activity | 1-phosphatidylinositol 4-kinase | F:protein binding | view details | ||
| JG646254.1 | Tinospora cordifolia | cytochrome P450 82C4-like | 638 | F:GO:0016491; F:GO:0046872; C:GO:0016020 | F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649057.1 | Tinospora cordifolia | Serine/threonine-protein kinase Aurora-1 | 712 | P:GO:0007052; P:GO:0032465; P:GO:0035404; F:GO:0004712; F:GO:0005524; F:GO:0035174; F:GO:0106310; C:GO:0005876; C:GO:0032133; C:GO:0051233 | P:mitotic spindle organization; P:regulation of cytokinesis; P:histone-serine phosphorylation; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:histone serine kinase activity; F:protein serine kinase activity; C:spindle microtubule; C:chromosome passenger complex; C:spindle midzone | Dual-specificity kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG649930.1 | Tinospora cordifolia | Structure of haze forming proteins in white wines: Vitis vinifera thaumatin-like proteins | 141 | P:GO:0006952 | P:defense response | no GO terms | view details | |||
| JG649698.1 | Tinospora cordifolia | probable pectin methyltransferase QUA2 | 632 | P:GO:0032259; F:GO:0008168; C:GO:0005737; C:GO:0016021; C:GO:0043231 | P:methylation; F:methyltransferase activity; C:cytoplasm; C:integral component of membrane; C:intracellular membrane-bounded organelle | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG648066.1 | Tinospora cordifolia | ATP synthase subunit beta, mitochondrial | 761 | P:GO:0042776; P:GO:1902600; F:GO:0005524; F:GO:0016887; F:GO:0046933; F:GO:0046961; C:GO:0000275; C:GO:0009536 | P:proton motive force-driven mitochondrial ATP synthesis; P:proton transmembrane transport; F:ATP binding; F:ATP hydrolysis activity; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:mitochondrial proton-transporting ATP synthase complex, catalytic sector F(1); C:plastid | H(+)-exporting diphosphatase; H(+)-transporting two-sector ATPase; Acting on acid anhydrides; Ligases | P:ATP biosynthetic process; P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding; F:ATP hydrolysis activity; C:mitochondrial proton-transporting ATP synthase complex, catalytic sector F(1) | view details | ||
| JG649203.1 | Tinospora cordifolia | basic endochitinase-like | 781 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646667.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At2g31400, chloroplastic-like | 779 | F:protein binding | view details | |||||
| JG650568.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 768 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648427.1 | Tinospora cordifolia | pyrophosphate--fructose 6-phosphate 1-phosphotransferase subunit alpha | 792 | P:GO:0006002; P:GO:0009749; P:GO:0015979; P:GO:0046835; P:GO:0061615; F:GO:0003872; F:GO:0005524; F:GO:0046872; F:GO:0047334; C:GO:0005829 | P:fructose 6-phosphate metabolic process; P:response to glucose; P:photosynthesis; P:carbohydrate phosphorylation; P:glycolytic process through fructose-6-phosphate; F:6-phosphofructokinase activity; F:ATP binding; F:metal ion binding; F:diphosphate-fructose-6-phosphate 1-phosphotransferase activity; C:cytosol | Diphosphate--fructose-6-phosphate 1-phosphotransferase; 6-phosphofructokinase | P:glycolytic process; F:6-phosphofructokinase activity | view details | ||
| JG646887.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 640 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649957.1 | Tinospora cordifolia | Zinc finger bed domain-containing protein daysleeper | 399 | P:GO:0009791; F:GO:0003677; F:GO:0046872; F:GO:0046983; C:GO:0005634 | P:post-embryonic development; F:DNA binding; F:metal ion binding; F:protein dimerization activity; C:nucleus | no GO terms | view details | |||
| JG648350.1 | Tinospora cordifolia | basic endochitinase-like | 858 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649395.1 | Tinospora cordifolia | bark storage protein A | 693 | P:GO:0009116; F:GO:0003824; C:GO:0016021 | P:nucleoside metabolic process; F:catalytic activity; C:integral component of membrane | P:nucleoside metabolic process; F:catalytic activity | view details | |||
| JG646647.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 864 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649663.1 | Tinospora cordifolia | probable carboxylesterase 12 | 790 | F:GO:0016787 | F:hydrolase activity | Hydrolases | F:hydrolase activity | view details | ||
| JG650356.1 | Tinospora cordifolia | protein TOPLESS-RELATED PROTEIN 2-like isoform X3 | 737 | P:GO:0006355; P:GO:0009791; P:GO:0048367; P:GO:0048608 | P:regulation of transcription, DNA-templated; P:post-embryonic development; P:shoot system development; P:reproductive structure development | P:regulation of transcription, DNA-templated; F:protein binding | view details | |||
| JG646425.1 | Tinospora cordifolia | pectinesterase 2 | 835 | P:GO:0042545; P:GO:0043086; P:GO:0045490; F:GO:0030599; F:GO:0045330; F:GO:0046910; C:GO:0005576 | P:cell wall modification; P:negative regulation of catalytic activity; P:pectin catabolic process; F:pectinesterase activity; F:aspartyl esterase activity; F:pectinesterase inhibitor activity; C:extracellular region | Pectinesterase | P:cell wall modification; F:enzyme inhibitor activity; F:pectinesterase activity | view details | ||
| JG646850.1 | Tinospora cordifolia | probable glutathione S-transferase | 621 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG649733.1 | Tinospora cordifolia | Glucan endo-1,3-beta-glucosidase | 584 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649610.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase 3-like isoform X1 | 816 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG649371.1 | Tinospora cordifolia | Corytuberine synthase | 719 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0005783; C:GO:0016020 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:endoplasmic reticulum; C:membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646804.1 | Tinospora cordifolia | tetraspanin-3-like | 820 | P:GO:0009734; C:GO:0009506; C:GO:0016021 | P:auxin-activated signaling pathway; C:plasmodesma; C:integral component of membrane | P:auxin-activated signaling pathway; C:integral component of membrane | view details | |||
| JG647141.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 786 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG645751.1 | Tinospora cordifolia | protein NRT1/ PTR FAMILY 5.10-like | 746 | P:GO:0006817; P:GO:0035442; P:GO:0042939; P:GO:0050896; F:GO:0042937; F:GO:0071916; C:GO:0016021 | P:phosphate ion transport; P:dipeptide transmembrane transport; P:tripeptide transport; P:response to stimulus; F:tripeptide transmembrane transporter activity; F:dipeptide transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| JG646093.1 | Tinospora cordifolia | beta-glucosidase BoGH3B-like | 644 | P:GO:0009251; F:GO:0008422; C:GO:0005576; C:GO:0016021 | P:glucan catabolic process; F:beta-glucosidase activity; C:extracellular region; C:integral component of membrane | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650556.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 721 | P:GO:0009058; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649349.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At1g08070, chloroplastic-like | 748 | F:protein binding | view details | |||||
| JG648093.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 884 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646609.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 66 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | no IPS match | view details | ||
| JG649727.1 | Tinospora cordifolia | ABC transporter-like | 624 | P:GO:0055085; F:GO:0005524; F:GO:0140359; C:GO:0016021 | P:transmembrane transport; F:ATP binding; F:ABC-type transporter activity; C:integral component of membrane | Translocases | C:membrane | view details | ||
| JG649681.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 678 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647798.1 | Tinospora cordifolia | probable aldo-keto reductase 2 | 850 | F:GO:0004033; C:GO:0005737 | F:aldo-keto reductase (NADP) activity; C:cytoplasm | Acting on the CH-OH group of donors | no GO terms | view details | ||
| LY749002.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 960 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG646436.1 | Tinospora cordifolia | ETHYLENE INSENSITIVE 3-like 1 protein | 833 | P:GO:0006355; P:GO:0009873; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:ethylene-activated signaling pathway; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | F:DNA-binding transcription factor activity; C:nucleus | view details | |||
| JG646365.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase 2-like | 834 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG646960.1 | Tinospora cordifolia | ADP-ribosylation factor GTPase-activating protein AGD4-like isoform X2 | 116 | P:GO:0050790; F:GO:0005096; F:GO:0046872; C:GO:0005737; C:GO:0016020 | P:regulation of catalytic activity; F:GTPase activator activity; F:metal ion binding; C:cytoplasm; C:membrane | no IPS match | view details | |||
| JG646824.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 747 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG646266.1 | Tinospora cordifolia | basic endochitinase-like | 691 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646019.1 | Tinospora cordifolia | CBL-interacting serine/threonine-protein kinase 14 | 782 | P:GO:0006468; P:GO:0007165; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310 | P:protein phosphorylation; P:signal transduction; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity | Dual-specificity kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG647482.1 | Tinospora cordifolia | cytochrome P450 81E8-like | 753 | F:GO:0016491 | F:oxidoreductase activity | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650209.1 | Tinospora cordifolia | indole-3-acetic acid-amido synthetase GH3.6 | 672 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG647672.1 | Tinospora cordifolia | S-adenosylmethionine synthetase | 193 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005737 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytoplasm | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG649697.1 | Tinospora cordifolia | ATP-dependent zinc metalloprotease FTSH 2, chloroplastic | 751 | P:GO:0006508; F:GO:0004176; F:GO:0004222; F:GO:0005524; F:GO:0008270; F:GO:0016887; C:GO:0009526; C:GO:0009534; C:GO:0016021 | P:proteolysis; F:ATP-dependent peptidase activity; F:metalloendopeptidase activity; F:ATP binding; F:zinc ion binding; F:ATP hydrolysis activity; C:plastid envelope; C:chloroplast thylakoid; C:integral component of membrane | Acting on peptide bonds (peptidases); Acting on acid anhydrides | no GO terms | view details | ||
| JG645697.1 | Tinospora cordifolia | isocitrate dehydrogenase [NAD] catalytic subunit 5, mitochondrial-like | 738 | P:GO:0006099; P:GO:0006102; F:GO:0000287; F:GO:0004449; F:GO:0051287; C:GO:0005739 | P:tricarboxylic acid cycle; P:isocitrate metabolic process; F:magnesium ion binding; F:isocitrate dehydrogenase (NAD+) activity; F:NAD binding; C:mitochondrion | Isocitrate dehydrogenase (NAD(+)) | no GO terms | view details | ||
| JG647471.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase 4 | 123 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG648161.1 | Tinospora cordifolia | centromere/kinetochore protein zw10 homolog | 826 | P:GO:0006888; P:GO:0007094; P:GO:0051301; C:GO:0005634; C:GO:0005737; C:GO:1990423 | P:endoplasmic reticulum to Golgi vesicle-mediated transport; P:mitotic spindle assembly checkpoint signaling; P:cell division; C:nucleus; C:cytoplasm; C:RZZ complex | P:mitotic cell cycle; C:chromosome, centromeric region; C:nucleus | view details | |||
| JG648238.1 | Tinospora cordifolia | Hsp70 family protein | 431 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG649573.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 833 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG645767.1 | Tinospora cordifolia | glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic | 858 | P:GO:0006006; P:GO:0006096; F:GO:0004365; F:GO:0050661; F:GO:0051287; C:GO:0005829; C:GO:0009536 | P:glucose metabolic process; P:glycolytic process; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity; F:NADP binding; F:NAD binding; C:cytosol; C:plastid | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating); Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG645676.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 821 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645543.1 | Tinospora cordifolia | ketol-acid reductoisomerase, chloroplastic | 673 | P:GO:0009097; P:GO:0009099; F:GO:0004455; F:GO:0016853; F:GO:0046872 | P:isoleucine biosynthetic process; P:valine biosynthetic process; F:ketol-acid reductoisomerase activity; F:isomerase activity; F:metal ion binding | Ketol-acid reductoisomerase (NADP(+)); Isomerases | P:branched-chain amino acid biosynthetic process; F:oxidoreductase activity | view details | ||
| JG650388.1 | Tinospora cordifolia | basic endochitinase-like | 842 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650176.1 | Tinospora cordifolia | 7-deoxyloganetin glucosyltransferase-like | 608 | P:GO:0009718; F:GO:0047213; F:GO:0080043; F:GO:0080044 | P:anthocyanin-containing compound biosynthetic process; F:anthocyanidin 3-O-glucosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Anthocyanidin 3-O-glucosyltransferase | no GO terms | view details | ||
| JG646125.1 | Tinospora cordifolia | S-adenosylmethionine synthase 1 | 751 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005737 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytoplasm | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG647248.1 | Tinospora cordifolia | Macpf domain-containing protein cad1 | 824 | P:GO:0009626; P:GO:2000031 | P:plant-type hypersensitive response; P:regulation of salicylic acid mediated signaling pathway | P:defense response; P:programmed cell death; P:regulation of salicylic acid mediated signaling pathway | view details | |||
| JG649313.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 755 | no GO terms | view details | |||||
| JG648094.1 | Tinospora cordifolia | basic endochitinase-like | 838 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647619.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 788 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648618.1 | Tinospora cordifolia | Multicopper oxidase | 878 | P:GO:0046274; F:GO:0005507; F:GO:0052716; C:GO:0048046 | P:lignin catabolic process; F:copper ion binding; F:hydroquinone:oxygen oxidoreductase activity; C:apoplast | Laccase | F:copper ion binding | view details | ||
| JG646141.1 | Tinospora cordifolia | 26S proteasome regulatory subunit 6A homolog | 772 | P:GO:0043161; P:GO:1901800; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0036402; C:GO:0005634; C:GO:0005737; C:GO:0008540 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; P:positive regulation of proteasomal protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:proteasome-activating activity; C:nucleus; C:cytoplasm; C:proteasome regulatory particle, base subcomplex | Acting on acid anhydrides; Acting on peptide bonds (peptidases); Proteasome ATPase | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG646571.1 | Tinospora cordifolia | FAD linked oxidase | 603 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646267.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 757 | P:GO:0005975; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649214.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 759 | P:GO:0009733; F:GO:0016881; C:GO:0005737; C:GO:0016021 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm; C:integral component of membrane | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG645820.1 | Tinospora cordifolia | 60S ribosomal protein L5 | 735 | P:GO:0000027; P:GO:0006412; F:GO:0003735; F:GO:0008097; C:GO:0005634; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:structural constituent of ribosome; F:5S rRNA binding; C:nucleus; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; F:5S rRNA binding; C:ribosome | view details | |||
| JG650346.1 | Tinospora cordifolia | protein MEI2-like 4 isoform X1 | 801 | F:GO:0003723 | F:RNA binding | F:nucleic acid binding; F:RNA binding | view details | |||
| JG647808.1 | Tinospora cordifolia | Corytuberine synthase | 748 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646481.1 | Tinospora cordifolia | (RS)-norcoclaurine 6-O-methyltransferase | 854 | P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757 | P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG646580.1 | Tinospora cordifolia | aldehyde dehydrogenase family 2 member B7, mitochondrial | 732 | F:GO:0004029; F:GO:0043878 | F:aldehyde dehydrogenase (NAD+) activity; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity | Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG646275.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 142 | P:GO:0005975; P:GO:0006952; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | no IPS match | view details | ||
| JG647801.1 | Tinospora cordifolia | long chain base biosynthesis protein 2a | 787 | P:GO:0046512; P:GO:0046513; F:GO:0004758; F:GO:0008483; F:GO:0030170; C:GO:0016021; C:GO:0017059 | P:sphingosine biosynthetic process; P:ceramide biosynthetic process; F:serine C-palmitoyltransferase activity; F:transaminase activity; F:pyridoxal phosphate binding; C:integral component of membrane; C:serine C-palmitoyltransferase complex | Transferring nitrogenous groups; Serine C-palmitoyltransferase | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG649550.1 | Tinospora cordifolia | long chain base biosynthesis protein 2a | 727 | P:GO:0046512; P:GO:0046513; F:GO:0004758; F:GO:0008483; F:GO:0030170; C:GO:0016021; C:GO:0017059 | P:sphingosine biosynthetic process; P:ceramide biosynthetic process; F:serine C-palmitoyltransferase activity; F:transaminase activity; F:pyridoxal phosphate binding; C:integral component of membrane; C:serine C-palmitoyltransferase complex | Transferring nitrogenous groups; Serine C-palmitoyltransferase | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG645808.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 820 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648709.1 | Tinospora cordifolia | UDP-glucuronic acid decarboxylase 5 | 835 | P:GO:0033320; P:GO:0042732; F:GO:0048040; F:GO:0070403; C:GO:0005737 | P:UDP-D-xylose biosynthetic process; P:D-xylose metabolic process; F:UDP-glucuronate decarboxylase activity; F:NAD+ binding; C:cytoplasm | UDP-glucuronate decarboxylase | P:D-xylose metabolic process; F:UDP-glucuronate decarboxylase activity; F:NAD+ binding | view details | ||
| JG647536.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 760 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG649008.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3-like | 459 | P:GO:0006913; P:GO:0015031; F:GO:0003924; F:GO:0005525; C:GO:0005634 | P:nucleocytoplasmic transport; P:protein transport; F:GTPase activity; F:GTP binding; C:nucleus | Acting on acid anhydrides | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG650170.1 | Tinospora cordifolia | berberine bridge enzyme | 497 | P:GO:0009820; F:GO:0050328; F:GO:0050468; F:GO:0071949; C:GO:0031410 | P:alkaloid metabolic process; F:tetrahydroberberine oxidase activity; F:reticuline oxidase activity; F:FAD binding; C:cytoplasmic vesicle | Reticuline oxidase; Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646221.1 | Tinospora cordifolia | pathogenesis-related protein 1 | 493 | P:GO:0006952; P:GO:0009607; C:GO:0005615 | P:defense response; P:response to biotic stimulus; C:extracellular space | no GO terms | view details | |||
| JG647331.1 | Tinospora cordifolia | Enoyl-CoA hydratase 2, peroxisomal | 788 | P:GO:0033542; F:GO:0003857; F:GO:0004300; F:GO:0044594; F:GO:0080023; C:GO:0005777 | P:fatty acid beta-oxidation, unsaturated, even number; F:3-hydroxyacyl-CoA dehydrogenase activity; F:enoyl-CoA hydratase activity; F:17-beta-hydroxysteroid dehydrogenase (NAD+) activity; F:3R-hydroxyacyl-CoA dehydratase activity; C:peroxisome | 3-hydroxyacyl-CoA dehydrogenase; 17-beta-estradiol 17-dehydrogenase; Enoyl-CoA hydratase | no GO terms | view details | ||
| JG647295.1 | Tinospora cordifolia | aspartate aminotransferase, cytoplasmic | 739 | P:GO:0006520; P:GO:1901566; F:GO:0004069; F:GO:0030170; C:GO:0005739 | P:cellular amino acid metabolic process; P:organonitrogen compound biosynthetic process; F:L-aspartate:2-oxoglutarate aminotransferase activity; F:pyridoxal phosphate binding; C:mitochondrion | Aspartate transaminase | P:cellular amino acid metabolic process; P:biosynthetic process; F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG648740.1 | Tinospora cordifolia | probable serine/threonine-protein kinase At1g54610 | 694 | P:GO:0032968; P:GO:0051726; P:GO:0070816; F:GO:0004693; F:GO:0005524; F:GO:0008353; C:GO:0000307; C:GO:0005634 | P:positive regulation of transcription elongation from RNA polymerase II promoter; P:regulation of cell cycle; P:phosphorylation of RNA polymerase II C-terminal domain; F:cyclin-dependent protein serine/threonine kinase activity; F:ATP binding; F:RNA polymerase II CTD heptapeptide repeat kinase activity; C:cyclin-dependent protein kinase holoenzyme complex; C:nucleus | [RNA-polymerase]-subunit kinase; Cyclin-dependent kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG650918.1 | Tinospora cordifolia | Omega-hydroxypalmitate O-feruloyl transferase | 780 | F:GO:0016747 | F:acyltransferase activity, transferring groups other than amino-acyl groups | Acyltransferases | no GO terms | view details | ||
| LY340439.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1569 | P:GO:0006259; P:GO:0006974; P:GO:0009058; P:GO:0051276; P:GO:0098542; F:GO:0000166; F:GO:0004386; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:DNA metabolic process; P:cellular response to DNA damage stimulus; P:biosynthetic process; P:chromosome organization; P:defense response to other organism; F:nucleotide binding; F:helicase activity; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647934.1 | Tinospora cordifolia | FRIGIDA-like protein 3 | 570 | P:GO:0009908; P:GO:0030154 | P:flower development; P:cell differentiation | no GO terms | view details | |||
| JG647286.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 789 | no GO terms | view details | |||||
| LY340442.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 1596 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650250.1 | Tinospora cordifolia | glutathione gamma-glutamylcysteinyltransferase 1-like isoform X1 | 730 | P:GO:0010273; P:GO:0046938; P:GO:0098849; F:GO:0016756; F:GO:0046872 | P:detoxification of copper ion; P:phytochelatin biosynthetic process; P:cellular detoxification of cadmium ion; F:glutathione gamma-glutamylcysteinyltransferase activity; F:metal ion binding | Glutathione gamma-glutamylcysteinyltransferase | P:response to metal ion; P:phytochelatin biosynthetic process; F:glutathione gamma-glutamylcysteinyltransferase activity; F:metal ion binding | view details | ||
| JG648678.1 | Tinospora cordifolia | BURP domain protein RD22-like | 847 | no GO terms | view details | |||||
| JG649977.1 | Tinospora cordifolia | CBL-interacting serine/threonine-protein kinase 6-like | 406 | P:GO:0006468; P:GO:0007165; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310 | P:protein phosphorylation; P:signal transduction; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity | Dual-specificity kinase | P:signal transduction | view details | ||
| JG648489.1 | Tinospora cordifolia | E3 ubiquitin-protein ligase COP1 | 819 | P:GO:0016567; P:GO:0043161; F:GO:0046872; F:GO:0061630 | P:protein ubiquitination; P:proteasome-mediated ubiquitin-dependent protein catabolic process; F:metal ion binding; F:ubiquitin protein ligase activity | Transferases | F:ubiquitin protein ligase activity | view details | ||
| JG650181.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 679 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG646117.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase | 819 | F:GO:0046872; F:GO:0051213 | F:metal ion binding; F:dioxygenase activity | Oxidoreductases | no GO terms | view details | ||
| JG649826.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 821 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650071.1 | Tinospora cordifolia | cullin-1 isoform X2 | 752 | P:GO:0006511; P:GO:0016567; F:GO:0008168; F:GO:0031625; C:GO:0016021; C:GO:0031461 | P:ubiquitin-dependent protein catabolic process; P:protein ubiquitination; F:methyltransferase activity; F:ubiquitin protein ligase binding; C:integral component of membrane; C:cullin-RING ubiquitin ligase complex | Transferring one-carbon groups | P:ubiquitin-dependent protein catabolic process; F:ubiquitin protein ligase binding | view details | ||
| LY749103.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 1596 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650589.1 | Tinospora cordifolia | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | 813 | P:GO:0032259; F:GO:0030776; F:GO:0030794; C:GO:0005737 | P:methylation; F:(RS)-1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase activity; F:(S)-coclaurine-N-methyltransferase activity; C:cytoplasm | (S)-coclaurine-N-methyltransferase; (RS)-1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase | no GO terms | view details | ||
| JG647947.1 | Tinospora cordifolia | serine carboxypeptidase II-2 | 702 | P:GO:0006508; F:GO:0004185; C:GO:0005576; C:GO:0016021 | P:proteolysis; F:serine-type carboxypeptidase activity; C:extracellular region; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type carboxypeptidase activity | view details | ||
| JG648957.1 | Tinospora cordifolia | monodehydroascorbate reductase | 632 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG650941.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 704 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649254.1 | Tinospora cordifolia | RS-norcoclaurine 6-O-methyltransferase-like protein | 726 | P:GO:0009821; P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757; F:GO:0042802; F:GO:0046983 | P:alkaloid biosynthetic process; P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; F:identical protein binding; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| JG647479.1 | Tinospora cordifolia | serine/threonine-protein phosphatase BSL3-like | 702 | P:GO:0006470; P:GO:0009742; F:GO:0017018; F:GO:0046872; C:GO:0005634 | P:protein dephosphorylation; P:brassinosteroid mediated signaling pathway; F:myosin phosphatase activity; F:metal ion binding; C:nucleus | Protein-serine/threonine phosphatase | F:hydrolase activity | view details | ||
| JG650956.1 | Tinospora cordifolia | ATP synthase subunit beta, mitochondrial | 831 | P:GO:0042776; P:GO:1902600; F:GO:0005524; F:GO:0016887; F:GO:0046933; F:GO:0046961; C:GO:0000275 | P:proton motive force-driven mitochondrial ATP synthesis; P:proton transmembrane transport; F:ATP binding; F:ATP hydrolysis activity; F:proton-transporting ATP synthase activity, rotational mechanism; F:proton-transporting ATPase activity, rotational mechanism; C:mitochondrial proton-transporting ATP synthase complex, catalytic sector F(1) | H(+)-exporting diphosphatase; H(+)-transporting two-sector ATPase; Acting on acid anhydrides; Ligases | P:proton motive force-driven ATP synthesis; F:ATP binding; F:proton-transporting ATP synthase activity, rotational mechanism; C:proton-transporting ATP synthase complex, catalytic core F(1) | view details | ||
| JG647705.1 | Tinospora cordifolia | NAD(P)H-quinone oxidoreductase subunit T, chloroplastic | 718 | C:GO:0009535; C:GO:0010598; C:GO:0016021 | C:chloroplast thylakoid membrane; C:NAD(P)H dehydrogenase complex (plastoquinone); C:integral component of membrane | no GO terms | view details | |||
| JG650432.1 | Tinospora cordifolia | dihydropyrimidinase isoform X2 | 718 | F:GO:0016810; C:GO:0005737 | F:hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds; C:cytoplasm | Acting on carbon-nitrogen bonds, other than peptide bonds | F:hydrolase activity | view details | ||
| JG649275.1 | Tinospora cordifolia | Sulfate transporter 3.1 | 798 | P:GO:1902358; F:GO:0008271; F:GO:0015301; C:GO:0005887; C:GO:0009507 | P:sulfate transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane; C:chloroplast | Translocases | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG649584.1 | Tinospora cordifolia | linamarin synthase 2-like | 778 | P:GO:0009718; F:GO:0047213; F:GO:0080043; F:GO:0080044 | P:anthocyanin-containing compound biosynthetic process; F:anthocyanidin 3-O-glucosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Anthocyanidin 3-O-glucosyltransferase | no GO terms | view details | ||
| JG646786.1 | Tinospora cordifolia | Multicopper oxidase | 748 | P:GO:0046274; F:GO:0005507; F:GO:0052716; C:GO:0048046 | P:lignin catabolic process; F:copper ion binding; F:hydroquinone:oxygen oxidoreductase activity; C:apoplast | Laccase | F:copper ion binding; F:oxidoreductase activity | view details | ||
| JG649912.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 162 | P:GO:0009416; F:GO:0016881; C:GO:0005737; C:GO:0016021 | P:response to light stimulus; F:acid-amino acid ligase activity; C:cytoplasm; C:integral component of membrane | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG646625.1 | Tinospora cordifolia | probable aquaporin PIP2-8 | 653 | P:GO:0006833; P:GO:0055085; F:GO:0015250; C:GO:0005886; C:GO:0016021 | P:water transport; P:transmembrane transport; F:water channel activity; C:plasma membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:channel activity; C:membrane | view details | ||
| JG648931.1 | Tinospora cordifolia | pathogenesis-related protein 1 | 100 | P:GO:0006952; P:GO:0009607; C:GO:0005576 | P:defense response; P:response to biotic stimulus; C:extracellular region | no IPS match | view details | |||
| JG646065.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 803 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG648404.1 | Tinospora cordifolia | fasciclin-like arabinogalactan protein 1 | 686 | C:GO:0016021; C:GO:0046658 | C:integral component of membrane; C:anchored component of plasma membrane | no GO terms | view details | |||
| JG648646.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 835 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648711.1 | Tinospora cordifolia | autophagy-related protein 2 isoform X1 | 851 | P:GO:0006914; C:GO:0005783; C:GO:0016020 | P:autophagy; C:endoplasmic reticulum; C:membrane | P:autophagy; P:autophagy of peroxisome | view details | |||
| JG648202.1 | Tinospora cordifolia | Tyrosine/DOPA decarboxylase | 831 | P:GO:0006520; F:GO:0004837; F:GO:0030170; C:GO:0005737; C:GO:0016021 | P:cellular amino acid metabolic process; F:tyrosine decarboxylase activity; F:pyridoxal phosphate binding; C:cytoplasm; C:integral component of membrane | Tyrosine decarboxylase | P:cellular amino acid metabolic process; P:carboxylic acid metabolic process; F:catalytic activity; F:carbon-carbon lyase activity; F:carboxy-lyase activity; F:pyridoxal phosphate binding | view details | ||
| JG649958.1 | Tinospora cordifolia | ran-binding protein M homolog | 197 | P:GO:0007010; P:GO:0007166; C:GO:0005737 | P:cytoskeleton organization; P:cell surface receptor signaling pathway; C:cytoplasm | no GO terms | view details | |||
| JG648001.1 | Tinospora cordifolia | diacylglycerol kinase 5 | 784 | P:GO:0006952; P:GO:0007205; P:GO:0016310; F:GO:0003951; F:GO:0004143; F:GO:0005524 | P:defense response; P:protein kinase C-activating G protein-coupled receptor signaling pathway; P:phosphorylation; F:NAD+ kinase activity; F:diacylglycerol kinase activity; F:ATP binding | NAD(+) kinase; Diacylglycerol kinase (ATP) | P:signal transduction; F:NAD+ kinase activity; F:diacylglycerol kinase activity; F:kinase activity | view details | ||
| JG649125.1 | Tinospora cordifolia | aromatic aminotransferase ISS1 | 800 | P:GO:0006555; P:GO:0006558; P:GO:0006569; P:GO:0006570; P:GO:0009641; P:GO:0009698; P:GO:0010252; P:GO:0010366; P:GO:1901997; F:GO:0010326; F:GO:0030170; C:GO:0005737 | P:methionine metabolic process; P:L-phenylalanine metabolic process; P:tryptophan catabolic process; P:tyrosine metabolic process; P:shade avoidance; P:phenylpropanoid metabolic process; P:auxin homeostasis; P:negative regulation of ethylene biosynthetic process; P:negative regulation of indoleacetic acid biosynthetic process via tryptophan; F:methionine-oxo-acid transaminase activity; F:pyridoxal phosphate binding; C:cytoplasm | Transferring nitrogenous groups | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG648914.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 764 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649104.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 787 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648544.1 | Tinospora cordifolia | 60S ribosomal protein L8 | 733 | P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649120.1 | Tinospora cordifolia | putative methyltransferase DDB_G0268948 | 707 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG645883.1 | Tinospora cordifolia | monodehydroascorbate reductase | 788 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG647159.1 | Tinospora cordifolia | pyruvate kinase, cytosolic isozyme | 774 | P:GO:0006096; F:GO:0000287; F:GO:0004743; F:GO:0005524; F:GO:0016301; F:GO:0030955; C:GO:0005737 | P:glycolytic process; F:magnesium ion binding; F:pyruvate kinase activity; F:ATP binding; F:kinase activity; F:potassium ion binding; C:cytoplasm | Pyruvate kinase | P:glycolytic process; F:magnesium ion binding; F:catalytic activity; F:pyruvate kinase activity; F:potassium ion binding | view details | ||
| JG647220.1 | Tinospora cordifolia | basic endochitinase-like | 840 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646649.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 729 | P:GO:0005975; F:GO:0004497; F:GO:0016705; F:GO:0016765; F:GO:0042973; F:GO:0046872; C:GO:0046658 | P:carbohydrate metabolic process; F:monooxygenase activity; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; F:metal ion binding; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647019.1 | Tinospora cordifolia | probable protein phosphatase 2C 23 | 791 | P:GO:0006470; F:GO:0016791; C:GO:0016020 | P:protein dephosphorylation; F:phosphatase activity; C:membrane | Acting on ester bonds | P:protein dephosphorylation; F:protein serine/threonine phosphatase activity | view details | ||
| JG649814.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 768 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646957.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 599 | no GO terms | view details | |||||
| JG649443.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like | 827 | F:GO:0016491; F:GO:0046872 | F:oxidoreductase activity; F:metal ion binding | Oxidoreductases | no GO terms | view details | ||
| JG645884.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 809 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646038.1 | Tinospora cordifolia | basic endochitinase-like | 836 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650694.1 | Tinospora cordifolia | basic endochitinase-like | 812 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646628.1 | Tinospora cordifolia | lysM domain receptor-like kinase 3 | 788 | P:GO:0006468; P:GO:0045087; F:GO:0004674; F:GO:0005524; F:GO:0019199; C:GO:0005886; C:GO:0016021 | P:protein phosphorylation; P:innate immune response; F:protein serine/threonine kinase activity; F:ATP binding; F:transmembrane receptor protein kinase activity; C:plasma membrane; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; P:innate immune response; F:protein kinase activity; F:ATP binding; F:transmembrane receptor protein kinase activity | view details | ||
| JG646899.1 | Tinospora cordifolia | isocitrate lyase | 727 | P:GO:0006097; P:GO:0006099; F:GO:0004451; F:GO:0046872; C:GO:0009514 | P:glyoxylate cycle; P:tricarboxylic acid cycle; F:isocitrate lyase activity; F:metal ion binding; C:glyoxysome | Isocitrate lyase | P:carboxylic acid metabolic process; F:catalytic activity; F:isocitrate lyase activity | view details | ||
| JG646303.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 840 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG646909.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 633 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650180.1 | Tinospora cordifolia | basic endochitinase-like | 672 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648339.1 | Tinospora cordifolia | Corytuberine synthase | 827 | P:GO:0009058; P:GO:0071704; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:biosynthetic process; P:organic substance metabolic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646970.1 | Tinospora cordifolia | formate dehydrogenase, mitochondrial | 176 | P:GO:0042183; F:GO:0008863; F:GO:0016616; F:GO:0051287; C:GO:0005739; C:GO:0009326; C:GO:0009507 | P:formate catabolic process; F:formate dehydrogenase (NAD+) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding; C:mitochondrion; C:formate dehydrogenase complex; C:chloroplast | Acting on the CH-OH group of donors; Formate dehydrogenase; Acting on the aldehyde or oxo group of donors | no IPS match | view details | ||
| JG647455.1 | Tinospora cordifolia | ferredoxin--nitrite reductase, chloroplastic-like | 759 | F:GO:0020037; F:GO:0046872; F:GO:0048307; F:GO:0051539; C:GO:0009570 | F:heme binding; F:metal ion binding; F:ferredoxin-nitrite reductase activity; F:4 iron, 4 sulfur cluster binding; C:chloroplast stroma | Ferredoxin--nitrite reductase | F:oxidoreductase activity; F:heme binding; F:iron-sulfur cluster binding | view details | ||
| JG645561.1 | Tinospora cordifolia | mitogen-activated protein kinase 20 | 733 | P:GO:0000165; P:GO:0006468; F:GO:0004707; F:GO:0005524; C:GO:0005634; C:GO:0005737; C:GO:0016021 | P:MAPK cascade; P:protein phosphorylation; F:MAP kinase activity; F:ATP binding; C:nucleus; C:cytoplasm; C:integral component of membrane | Mitogen-activated protein kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG646906.1 | Tinospora cordifolia | sulfate transporter 3.1-like | 665 | P:GO:1902358; F:GO:0008271; F:GO:0015293; F:GO:0015301; C:GO:0005887; C:GO:0009507 | P:sulfate transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:symporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane; C:chloroplast | Translocases | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:chloroplast; C:membrane; C:integral component of membrane | view details | ||
| JG649146.1 | Tinospora cordifolia | Polygalacturonase inhibitor | 435 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG647490.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 772 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646505.1 | Tinospora cordifolia | CLP protease regulatory subunit CLPX1, mitochondrial-like | 789 | P:GO:0006457; P:GO:0006508; P:GO:0030163; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0051082; C:GO:0005759 | P:protein folding; P:proteolysis; P:protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:unfolded protein binding; C:mitochondrial matrix | Acting on acid anhydrides; Acting on peptide bonds (peptidases) | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG647231.1 | Tinospora cordifolia | 2-hydroxyacyl-CoA lyase | 827 | P:GO:0001561; F:GO:0000287; F:GO:0016829; F:GO:0030976; C:GO:0005777 | P:fatty acid alpha-oxidation; F:magnesium ion binding; F:lyase activity; F:thiamine pyrophosphate binding; C:peroxisome | Lyases | F:magnesium ion binding; F:thiamine pyrophosphate binding | view details | ||
| JG646268.1 | Tinospora cordifolia | root phototropism protein 2 | 712 | P:GO:0009638; P:GO:0016567 | P:phototropism; P:protein ubiquitination | P:phototropism; F:protein binding | view details | |||
| JG650661.1 | Tinospora cordifolia | sulfate transporter 3.1-like | 814 | P:GO:1902358; F:GO:0008271; F:GO:0015293; F:GO:0015301; C:GO:0005887; C:GO:0009507 | P:sulfate transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:symporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane; C:chloroplast | Translocases | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:chloroplast; C:membrane; C:integral component of membrane | view details | ||
| JG649420.1 | Tinospora cordifolia | BURP domain protein RD22-like | 769 | no GO terms | view details | |||||
| JG648586.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 768 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650318.1 | Tinospora cordifolia | glutamate--glyoxylate aminotransferase 2 | 824 | P:GO:0009853; P:GO:0042853; P:GO:1901566; F:GO:0004021; F:GO:0008453; F:GO:0030170; F:GO:0047958 | P:photorespiration; P:L-alanine catabolic process; P:organonitrogen compound biosynthetic process; F:L-alanine:2-oxoglutarate aminotransferase activity; F:alanine-glyoxylate transaminase activity; F:pyridoxal phosphate binding; F:glycine:2-oxoglutarate aminotransferase activity | Alanine--glyoxylate transaminase; Alanine--oxo-acid transaminase; Alanine transaminase | P:biosynthetic process; F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG649011.1 | Tinospora cordifolia | 4-coumarate--CoA ligase-like 5 | 128 | P:GO:0009611; P:GO:0009617; P:GO:0009695; P:GO:0009753; F:GO:0016405; C:GO:0005777; C:GO:0016021 | P:response to wounding; P:response to bacterium; P:jasmonic acid biosynthetic process; P:response to jasmonic acid; F:CoA-ligase activity; C:peroxisome; C:integral component of membrane | Forming carbon-sulfur bonds | no IPS match | view details | ||
| JG646797.1 | Tinospora cordifolia | mitochondrial-processing peptidase subunit alpha-like | 816 | P:GO:0006627; F:GO:0004222; F:GO:0046872; C:GO:0005743; C:GO:0005759 | P:protein processing involved in protein targeting to mitochondrion; F:metalloendopeptidase activity; F:metal ion binding; C:mitochondrial inner membrane; C:mitochondrial matrix | Acting on peptide bonds (peptidases) | P:protein processing involved in protein targeting to mitochondrion; F:metal ion binding | view details | ||
| JG650172.1 | Tinospora cordifolia | ---NA--- | 73 | no IPS match | view details | |||||
| JG646892.1 | Tinospora cordifolia | Glucan endo-1,3-beta-glucosidase | 586 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647221.1 | Tinospora cordifolia | triosephosphate isomerase, cytosolic | 773 | P:GO:0006094; P:GO:0006096; P:GO:0019563; P:GO:0046166; F:GO:0004807; C:GO:0005829 | P:gluconeogenesis; P:glycolytic process; P:glycerol catabolic process; P:glyceraldehyde-3-phosphate biosynthetic process; F:triose-phosphate isomerase activity; C:cytosol | Triose-phosphate isomerase | P:glycolytic process; F:triose-phosphate isomerase activity | view details | ||
| JG645896.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 781 | F:GO:0050660 | F:flavin adenine dinucleotide binding | F:flavin adenine dinucleotide binding | view details | |||
| JG650946.1 | Tinospora cordifolia | cyclic dof factor 2-like | 802 | P:GO:0006355; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity | view details | |||
| JG646865.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase | 659 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0016021 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:integral component of membrane | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG649786.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 847 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG648316.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 725 | no GO terms | view details | |||||
| JG649736.1 | Tinospora cordifolia | autophagy-related protein 18g-like isoform X1 | 719 | P:GO:0006914; P:GO:0042594; F:GO:0016740; C:GO:0005737 | P:autophagy; P:response to starvation; F:transferase activity; C:cytoplasm | Transferases | P:autophagy | view details | ||
| JG646720.1 | Tinospora cordifolia | cullin-4 | 846 | P:GO:0006511; P:GO:0016567; F:GO:0008168; F:GO:0030674; F:GO:0031625; C:GO:0031461 | P:ubiquitin-dependent protein catabolic process; P:protein ubiquitination; F:methyltransferase activity; F:protein-macromolecule adaptor activity; F:ubiquitin protein ligase binding; C:cullin-RING ubiquitin ligase complex | Transferring one-carbon groups | P:ubiquitin-dependent protein catabolic process; F:ubiquitin protein ligase binding | view details | ||
| JG649641.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 786 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648313.1 | Tinospora cordifolia | polyubiquitin 3 | 745 | P:GO:0006511; P:GO:0010224; P:GO:0016567; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:ubiquitin-dependent protein catabolic process; P:response to UV-B; P:protein ubiquitination; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | F:protein binding | view details | |||
| JG650641.1 | Tinospora cordifolia | oxalate--CoA ligase | 866 | P:GO:0006631; P:GO:0009698; P:GO:0010030; P:GO:0010214; P:GO:0033611; P:GO:0050832; F:GO:0016207; F:GO:0031956; F:GO:0050203; C:GO:0005737 | P:fatty acid metabolic process; P:phenylpropanoid metabolic process; P:positive regulation of seed germination; P:seed coat development; P:oxalate catabolic process; P:defense response to fungus; F:4-coumarate-CoA ligase activity; F:medium-chain fatty acid-CoA ligase activity; F:oxalate-CoA ligase activity; C:cytoplasm | 4-coumarate--CoA ligase; Oxalate--CoA ligase; Medium-chain acyl-CoA ligase | no GO terms | view details | ||
| JG647297.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 698 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650830.1 | Tinospora cordifolia | polyubiquitin 11 | 689 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG648398.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP) | 700 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG647215.1 | Tinospora cordifolia | Luminal-binding protein 5 | 795 | P:GO:0030433; P:GO:0030968; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005634; C:GO:0005788; C:GO:0016020; C:GO:0034663 | P:ubiquitin-dependent ERAD pathway; P:endoplasmic reticulum unfolded protein response; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:nucleus; C:endoplasmic reticulum lumen; C:membrane; C:endoplasmic reticulum chaperone complex | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG647151.1 | Tinospora cordifolia | Uridine kinase | 747 | P:GO:0016310; F:GO:0005524; F:GO:0016301; F:GO:0016787; C:GO:0009507; C:GO:0016021 | P:phosphorylation; F:ATP binding; F:kinase activity; F:hydrolase activity; C:chloroplast; C:integral component of membrane | Transferring phosphorus-containing groups; Hydrolases | F:ATP binding; F:kinase activity | view details | ||
| JG648944.1 | Tinospora cordifolia | uncharacterized mitochondrial protein AtMg00810-like | 726 | no GO terms | view details | |||||
| JG650038.1 | Tinospora cordifolia | ADP,ATP carrier protein, mitochondrial-like | 788 | P:GO:0140021; P:GO:1990544; F:GO:0005471; C:GO:0005743; C:GO:0016021 | P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane; C:integral component of membrane | Translocases | P:transmembrane transport; P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane | view details | ||
| JG648499.1 | Tinospora cordifolia | serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform isoform X1 | 637 | P:GO:0006811; P:GO:0015986; P:GO:0050790; P:GO:0070262; F:GO:0015078; F:GO:0019888; C:GO:0000159; C:GO:0000276; C:GO:0005829 | P:ion transport; P:proton motive force-driven ATP synthesis; P:regulation of catalytic activity; P:peptidyl-serine dephosphorylation; F:proton transmembrane transporter activity; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex; C:mitochondrial proton-transporting ATP synthase complex, coupling factor F(o); C:cytosol | Translocases | F:protein binding; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex | view details | ||
| JG646230.1 | Tinospora cordifolia | probable serine acetyltransferase 1 | 101 | P:GO:0006535; F:GO:0009001; C:GO:0005829 | P:cysteine biosynthetic process from serine; F:serine O-acetyltransferase activity; C:cytosol | Serine O-acetyltransferase | no IPS match | view details | ||
| JG645855.1 | Tinospora cordifolia | ABC transporter I family member 19 | 790 | F:GO:0000166; F:GO:0005524 | F:nucleotide binding; F:ATP binding | F:ATP binding | view details | |||
| JG647783.1 | Tinospora cordifolia | protein SEMI-ROLLED LEAF 2 isoform X2 | 758 | no GO terms | view details | |||||
| JG649277.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 815 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650416.1 | Tinospora cordifolia | basic endochitinase | 783 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648898.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 691 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; P:methionine biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647015.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 742 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464; F:GO:0071949 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity; F:FAD binding | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG646942.1 | Tinospora cordifolia | Pathogenesis-related protein 1 | 441 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | no IPS match | view details | |||
| JG650488.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 804 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648728.1 | Tinospora cordifolia | probable methyltransferase PMT18 | 808 | P:GO:0032259; F:GO:0008168; C:GO:0005737; C:GO:0016021; C:GO:0043231 | P:methylation; F:methyltransferase activity; C:cytoplasm; C:integral component of membrane; C:intracellular membrane-bounded organelle | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG647276.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 718 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650651.1 | Tinospora cordifolia | 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 | 835 | P:GO:0008299; P:GO:0016126; F:GO:0004420; C:GO:0005778; C:GO:0005789 | P:isoprenoid biosynthetic process; P:sterol biosynthetic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; C:peroxisomal membrane; C:endoplasmic reticulum membrane | Hydroxymethylglutaryl-CoA reductase (NADPH) | P:coenzyme A metabolic process; F:hydroxymethylglutaryl-CoA reductase (NADPH) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG650230.1 | Tinospora cordifolia | fructose-bisphosphate aldolase 3, chloroplastic | 682 | P:GO:0006096; P:GO:0030388; F:GO:0004332; C:GO:0005829 | P:glycolytic process; P:fructose 1,6-bisphosphate metabolic process; F:fructose-bisphosphate aldolase activity; C:cytosol | Fructose-bisphosphate aldolase | P:glycolytic process; F:fructose-bisphosphate aldolase activity | view details | ||
| JG645971.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 884 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649459.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 859 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647098.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 779 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG650578.1 | Tinospora cordifolia | aspartic proteinase A1-like | 833 | P:GO:0006508; P:GO:0006629; F:GO:0004190; C:GO:0016021 | P:proteolysis; P:lipid metabolic process; F:aspartic-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; P:lipid metabolic process; P:sphingolipid metabolic process; F:aspartic-type endopeptidase activity; C:lysosome | view details | ||
| JG649952.1 | Tinospora cordifolia | monodehydroascorbate reductase | 456 | F:GO:0016656; F:GO:0050660; C:GO:0005737 | F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG645848.1 | Tinospora cordifolia | carbon catabolite repressor protein 4 homolog 1 | 814 | P:GO:0000289; P:GO:0090503; F:GO:0004519; F:GO:0004535; C:GO:0005634; C:GO:0005737; C:GO:0030014 | P:nuclear-transcribed mRNA poly(A) tail shortening; P:RNA phosphodiester bond hydrolysis, exonucleolytic; F:endonuclease activity; F:poly(A)-specific ribonuclease activity; C:nucleus; C:cytoplasm; C:CCR4-NOT complex | Acting on ester bonds; Poly(A)-specific ribonuclease | no GO terms | view details | ||
| JG646835.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 686 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647191.1 | Tinospora cordifolia | eukaryotic translation initiation factor 3 subunit A-like | 735 | P:GO:0001732; P:GO:0002188; F:GO:0003729; F:GO:0003743; C:GO:0000502; C:GO:0016021; C:GO:0016282; C:GO:0033290; C:GO:0043614; C:GO:0071540; C:GO:0071541 | P:formation of cytoplasmic translation initiation complex; P:translation reinitiation; F:mRNA binding; F:translation initiation factor activity; C:proteasome complex; C:integral component of membrane; C:eukaryotic 43S preinitiation complex; C:eukaryotic 48S preinitiation complex; C:multi-eIF complex; C:eukaryotic translation initiation factor 3 complex, eIF3e; C:eukaryotic translation initiation factor 3 complex, eIF3m | C:eukaryotic translation initiation factor 3 complex | view details | |||
| JG646891.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 725 | F:GO:0016881; C:GO:0005737; C:GO:0016021 | F:acid-amino acid ligase activity; C:cytoplasm; C:integral component of membrane | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG646160.1 | Tinospora cordifolia | basic endochitinase-like | 803 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| LY340341.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 960 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG649889.1 | Tinospora cordifolia | ribonuclease 3 | 290 | P:GO:0006401; P:GO:0090502; F:GO:0003723; F:GO:0033897; C:GO:0005576 | P:RNA catabolic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA binding; F:ribonuclease T2 activity; C:extracellular region | Acting on ester bonds; Ribonuclease T(2) | F:RNA binding; F:ribonuclease T2 activity | view details | ||
| JG646209.1 | Tinospora cordifolia | uroporphyrinogen-III C-methyltransferase | 643 | P:GO:0009416; P:GO:0019354; P:GO:0032259; P:GO:0090352; P:GO:1900058; P:GO:1902326; F:GO:0004851; C:GO:0009507 | P:response to light stimulus; P:siroheme biosynthetic process; P:methylation; P:regulation of nitrate assimilation; P:regulation of sulfate assimilation; P:positive regulation of chlorophyll biosynthetic process; F:uroporphyrin-III C-methyltransferase activity; C:chloroplast | Uroporphyrinogen-III C-methyltransferase | F:methyltransferase activity | view details | ||
| JG646963.1 | Tinospora cordifolia | (S)-norcoclaurine synthase | 318 | P:GO:0002238; P:GO:0006952; P:GO:1901010; F:GO:0050474; C:GO:0016021 | P:response to molecule of fungal origin; P:defense response; P:(S)-reticuline metabolic process; F:(S)-norcoclaurine synthase activity; C:integral component of membrane | (S)-norcoclaurine synthase | no GO terms | view details | ||
| JG646997.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 849 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG646094.1 | Tinospora cordifolia | cytochrome P450 81E8-like | 821 | P:GO:0009058; F:GO:0005488; F:GO:0016491 | P:biosynthetic process; F:binding; F:oxidoreductase activity | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650064.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 801 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649859.1 | Tinospora cordifolia | basic endochitinase | 762 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647767.1 | Tinospora cordifolia | ABC transporter I family member 19 | 786 | F:GO:0005524 | F:ATP binding | F:ATP binding | view details | |||
| JG646661.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 790 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648525.1 | Tinospora cordifolia | putative ribosome biogenesis protein C8F11.04 | 734 | C:GO:0005840 | C:ribosome | no GO terms | view details | |||
| JG646701.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 768 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG649562.1 | Tinospora cordifolia | caffeic acid 3-O-methyltransferase | 857 | P:GO:0009699; P:GO:0032259; F:GO:0008171; F:GO:0008757 | P:phenylpropanoid biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG646249.1 | Tinospora cordifolia | phytosulfokine receptor 1-like | 487 | P:GO:0016310; P:GO:0044238; P:GO:0098542; P:GO:1901564; F:GO:0000166; F:GO:0004672; C:GO:0016020 | P:phosphorylation; P:primary metabolic process; P:defense response to other organism; P:organonitrogen compound metabolic process; F:nucleotide binding; F:protein kinase activity; C:membrane | Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JG650957.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 791 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| LQ667396.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1617 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649622.1 | Tinospora cordifolia | monodehydroascorbate reductase | 798 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG649233.1 | Tinospora cordifolia | pathogen-related protein-like | 233 | no GO terms | view details | |||||
| JG647797.1 | Tinospora cordifolia | (-)-germacrene D synthase-like | 814 | P:GO:0016102; F:GO:0000287; F:GO:0010333 | P:diterpenoid biosynthetic process; F:magnesium ion binding; F:terpene synthase activity | Carbon-oxygen lyases | F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| JG648432.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 801 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648680.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 783 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464; F:GO:0071949 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity; F:FAD binding | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG650129.1 | Tinospora cordifolia | ADP,ATP carrier protein, mitochondrial | 742 | P:GO:0140021; P:GO:1990544; F:GO:0005471; C:GO:0005743; C:GO:0016021 | P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane; C:integral component of membrane | Translocases | P:transmembrane transport; P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane | view details | ||
| JG648495.1 | Tinospora cordifolia | 26S proteasome regulatory subunit 6B homolog | 764 | P:GO:0043161; P:GO:1901800; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0036402; C:GO:0005634; C:GO:0005737; C:GO:0008540 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; P:positive regulation of proteasomal protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:proteasome-activating activity; C:nucleus; C:cytoplasm; C:proteasome regulatory particle, base subcomplex | Acting on acid anhydrides; Acting on peptide bonds (peptidases); Proteasome ATPase | P:protein catabolic process; F:ATP binding; F:ATP hydrolysis activity; F:proteasome-activating activity; C:cytoplasm | view details | ||
| JG649685.1 | Tinospora cordifolia | acetyl-CoA acetyltransferase, cytosolic 1 | 780 | P:GO:0006635; P:GO:0009846; P:GO:0009860; P:GO:0016125; F:GO:0003985 | P:fatty acid beta-oxidation; P:pollen germination; P:pollen tube growth; P:sterol metabolic process; F:acetyl-CoA C-acetyltransferase activity | Acetyl-CoA C-acyltransferase; Acetyl-CoA C-acetyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG649888.1 | Tinospora cordifolia | type II peroxiredoxin C | 538 | P:GO:0034599; P:GO:0042744; P:GO:0045454; P:GO:0098869; F:GO:0008379; C:GO:0005737 | P:cellular response to oxidative stress; P:hydrogen peroxide catabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:thioredoxin peroxidase activity; C:cytoplasm | Thioredoxin-dependent peroxiredoxin | F:oxidoreductase activity | view details | ||
| JG647710.1 | Tinospora cordifolia | serine hydroxymethyltransferase 4 | 771 | P:GO:0006565; P:GO:0019264; P:GO:0032259; P:GO:0035999; P:GO:0046655; F:GO:0004372; F:GO:0008168; F:GO:0008270; F:GO:0030170; F:GO:0070905; C:GO:0005737 | P:L-serine catabolic process; P:glycine biosynthetic process from serine; P:methylation; P:tetrahydrofolate interconversion; P:folic acid metabolic process; F:glycine hydroxymethyltransferase activity; F:methyltransferase activity; F:zinc ion binding; F:pyridoxal phosphate binding; F:serine binding; C:cytoplasm | Glycine hydroxymethyltransferase; Transferring one-carbon groups | P:glycine biosynthetic process from serine; P:tetrahydrofolate interconversion; F:catalytic activity; F:glycine hydroxymethyltransferase activity; F:pyridoxal phosphate binding | view details | ||
| JG650760.1 | Tinospora cordifolia | class V chitinase-like | 814 | P:GO:0006032; F:GO:0004568; F:GO:0008061; C:GO:0005576 | P:chitin catabolic process; F:chitinase activity; F:chitin binding; C:extracellular region | Chitinase | P:carbohydrate metabolic process | view details | ||
| JG649407.1 | Tinospora cordifolia | oxysterol-binding protein-related protein 1C isoform X2 | 719 | P:GO:0015918; F:GO:0015248; F:GO:0032934; C:GO:0005829; C:GO:0016020; C:GO:0043231 | P:sterol transport; F:sterol transporter activity; F:sterol binding; C:cytosol; C:membrane; C:intracellular membrane-bounded organelle | Translocases | F:lipid binding | view details | ||
| JG647741.1 | Tinospora cordifolia | NF-X1-type zinc finger protein NFXL1-like | 746 | P:GO:0006357; P:GO:0006412; F:GO:0000977; F:GO:0000981; F:GO:0003723; F:GO:0003735; F:GO:0008270; C:GO:0005634; C:GO:0015935 | P:regulation of transcription by RNA polymerase II; P:translation; F:RNA polymerase II transcription regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity, RNA polymerase II-specific; F:RNA binding; F:structural constituent of ribosome; F:zinc ion binding; C:nucleus; C:small ribosomal subunit | F:DNA-binding transcription factor activity | view details | |||
| JG649213.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 698 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG646448.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 830 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650048.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 692 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646263.1 | Tinospora cordifolia | probable plastidic glucose transporter 2 isoform X1 | 746 | P:GO:0006817; P:GO:0008645; P:GO:0050896; F:GO:0015149; C:GO:0016021 | P:phosphate ion transport; P:hexose transmembrane transport; P:response to stimulus; F:hexose transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG648263.1 | Tinospora cordifolia | basic endochitinase | 710 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647502.1 | Tinospora cordifolia | uridine nucleosidase 1 | 787 | P:GO:0006152; F:GO:0008477; C:GO:0005829 | P:purine nucleoside catabolic process; F:purine nucleosidase activity; C:cytosol | Purine nucleosidase | F:hydrolase activity, hydrolyzing N-glycosyl compounds | view details | ||
| JG649582.1 | Tinospora cordifolia | UPF0051 protein ABCI8, chloroplastic | 832 | P:GO:0016226 | P:iron-sulfur cluster assembly | no GO terms | view details | |||
| JG646951.1 | Tinospora cordifolia | monodehydroascorbate reductase | 684 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG645535.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 834 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG646980.1 | Tinospora cordifolia | AT5G39320-like protein | 123 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0016628; F:GO:0051287; C:GO:0005634; C:GO:0005829; C:GO:0016021 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor; F:NAD binding; C:nucleus; C:cytosol; C:integral component of membrane | UDP-glucose 6-dehydrogenase; Acting on the CH-CH group of donors | no IPS match | view details | ||
| MS687777.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1581 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646286.1 | Tinospora cordifolia | V-type proton ATPase subunit B3 | 709 | P:GO:0046034; P:GO:1902600; F:GO:0005524; F:GO:0046961; C:GO:0009507; C:GO:0031090; C:GO:0033180 | P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding; F:proton-transporting ATPase activity, rotational mechanism; C:chloroplast; C:organelle membrane; C:proton-transporting V-type ATPase, V1 domain | H(+)-exporting diphosphatase | P:ATP metabolic process; P:proton transmembrane transport; F:ATP binding | view details | ||
| JG649600.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 758 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646706.1 | Tinospora cordifolia | phosphoglucomutase, cytoplasmic | 804 | P:GO:0006006; F:GO:0000287; F:GO:0004614; C:GO:0005829 | P:glucose metabolic process; F:magnesium ion binding; F:phosphoglucomutase activity; C:cytosol | Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | P:carbohydrate metabolic process; F:phosphoglucomutase activity; F:intramolecular transferase activity, phosphotransferases | view details | ||
| JG648474.1 | Tinospora cordifolia | basic endochitinase-like | 835 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG645874.1 | Tinospora cordifolia | dynamin-related protein 5A | 813 | F:GO:0003924; F:GO:0005525; F:GO:0008017; C:GO:0005737; C:GO:0005874; C:GO:0016020 | F:GTPase activity; F:GTP binding; F:microtubule binding; C:cytoplasm; C:microtubule; C:membrane | Acting on acid anhydrides | no GO terms | view details | ||
| JG647392.1 | Tinospora cordifolia | ---NA--- | 94 | no IPS match | view details | |||||
| JG646731.1 | Tinospora cordifolia | ADP,ATP carrier protein 3, mitochondrial-like | 825 | P:GO:0140021; P:GO:1990544; F:GO:0005471; C:GO:0005743; C:GO:0016021 | P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane; C:integral component of membrane | Translocases | P:transmembrane transport; P:mitochondrial ADP transmembrane transport; P:mitochondrial ATP transmembrane transport; F:ATP:ADP antiporter activity; C:mitochondrial inner membrane | view details | ||
| JG647277.1 | Tinospora cordifolia | serine hydroxymethyltransferase 7-like | 833 | P:GO:0006565; P:GO:0019264; P:GO:0032259; P:GO:0035999; P:GO:0046655; F:GO:0004372; F:GO:0008168; F:GO:0008270; F:GO:0030170; F:GO:0070905; C:GO:0005737 | P:L-serine catabolic process; P:glycine biosynthetic process from serine; P:methylation; P:tetrahydrofolate interconversion; P:folic acid metabolic process; F:glycine hydroxymethyltransferase activity; F:methyltransferase activity; F:zinc ion binding; F:pyridoxal phosphate binding; F:serine binding; C:cytoplasm | Glycine hydroxymethyltransferase; Transferring one-carbon groups | P:glycine biosynthetic process from serine; P:tetrahydrofolate interconversion; F:catalytic activity; F:glycine hydroxymethyltransferase activity; F:pyridoxal phosphate binding | view details | ||
| JG649973.1 | Tinospora cordifolia | basic endochitinase-like | 126 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649718.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 621 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG646301.1 | Tinospora cordifolia | probable phosphoribosylformylglycinamidine synthase, chloroplastic/mitochondrial | 772 | P:GO:0006189; P:GO:0006541; P:GO:0055046; F:GO:0004642; F:GO:0005524; F:GO:0046872; C:GO:0005739; C:GO:0009507 | P:de novo IMP biosynthetic process; P:glutamine metabolic process; P:microgametogenesis; F:phosphoribosylformylglycinamidine synthase activity; F:ATP binding; F:metal ion binding; C:mitochondrion; C:chloroplast | Phosphoribosylformylglycinamidine synthase | no GO terms | view details | ||
| JG650722.1 | Tinospora cordifolia | mogroside IE synthase-like | 704 | F:GO:0016757 | F:glycosyltransferase activity | Glycosyltransferases | no GO terms | view details | ||
| JG648783.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 870 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG647110.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 802 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG648458.1 | Tinospora cordifolia | Cyclin-dependent kinase f-4 | 804 | P:GO:0006468; P:GO:0035556; F:GO:0004674; F:GO:0005524; C:GO:0005634; C:GO:0005737 | P:protein phosphorylation; P:intracellular signal transduction; F:protein serine/threonine kinase activity; F:ATP binding; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG650087.1 | Tinospora cordifolia | serine carboxypeptidase II-2 | 810 | P:GO:0006508; F:GO:0004185; C:GO:0016020 | P:proteolysis; F:serine-type carboxypeptidase activity; C:membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type carboxypeptidase activity | view details | ||
| JG646735.1 | Tinospora cordifolia | basic endochitinase-like | 804 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650754.1 | Tinospora cordifolia | UDP-glycosyltransferase 74F2-like | 710 | F:GO:0080043; F:GO:0080044 | F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Glycosyltransferases | no GO terms | view details | ||
| JG647361.1 | Tinospora cordifolia | serine carboxypeptidase-like 29 | 457 | P:GO:0006508; F:GO:0004185; C:GO:0005576 | P:proteolysis; F:serine-type carboxypeptidase activity; C:extracellular region | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type carboxypeptidase activity | view details | ||
| MA258133.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 966 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG649037.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 554 | P:GO:0010951; F:GO:0004866; C:GO:0005576 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity; C:extracellular region | F:endopeptidase inhibitor activity | view details | |||
| JG648967.1 | Tinospora cordifolia | Glucan endo-1,3-beta-glucosidase | 244 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647739.1 | Tinospora cordifolia | replication protein A 32 kDa subunit A-like isoform X2 | 851 | P:GO:0000724; P:GO:0006260; P:GO:0006289; F:GO:0003697; F:GO:0016740; C:GO:0000781; C:GO:0005662; C:GO:0035861 | P:double-strand break repair via homologous recombination; P:DNA replication; P:nucleotide-excision repair; F:single-stranded DNA binding; F:transferase activity; C:chromosome, telomeric region; C:DNA replication factor A complex; C:site of double-strand break | Transferases | P:DNA replication; P:DNA repair; P:DNA recombination; F:DNA binding; C:nucleus | view details | ||
| JG648509.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 746 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646695.1 | Tinospora cordifolia | ferredoxin--nitrite reductase, chloroplastic-like | 823 | F:GO:0020037; F:GO:0046872; F:GO:0048307; F:GO:0051539; C:GO:0009570 | F:heme binding; F:metal ion binding; F:ferredoxin-nitrite reductase activity; F:4 iron, 4 sulfur cluster binding; C:chloroplast stroma | Ferredoxin--nitrite reductase | F:oxidoreductase activity; F:heme binding; F:iron-sulfur cluster binding | view details | ||
| JG648890.1 | Tinospora cordifolia | epidermis-specific secreted glycoprotein EP1-like | 800 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG648926.1 | Tinospora cordifolia | Indole-3-acetic acid-amido synthetase GH3.6 | 811 | F:GO:0010279; C:GO:0005737 | F:indole-3-acetic acid amido synthetase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| KT384097.1 | Tinospora cordifolia | pol protein | 547 | P:GO:0015074; F:GO:0003676 | P:DNA integration; F:nucleic acid binding | F:nucleic acid binding | view details | |||
| JG646959.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 207 | P:GO:0009416; F:GO:0016881; C:GO:0005737 | P:response to light stimulus; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no IPS match | view details | ||
| JG650942.1 | Tinospora cordifolia | serine/threonine-protein kinase TOUSLED isoform X1 | 768 | P:GO:0007059; P:GO:0018105; P:GO:0035556; F:GO:0004674; F:GO:0005524; C:GO:0005634 | P:chromosome segregation; P:peptidyl-serine phosphorylation; P:intracellular signal transduction; F:protein serine/threonine kinase activity; F:ATP binding; C:nucleus | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG649026.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 478 | P:GO:0005975; P:GO:0006952; F:GO:0005506; F:GO:0016712; F:GO:0016765; F:GO:0020037; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:heme binding; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648988.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 279 | no GO terms | view details | |||||
| JG649732.1 | Tinospora cordifolia | long chain acyl-CoA synthetase 4-like | 679 | P:GO:0001676; F:GO:0004467; F:GO:0005524; C:GO:0005783; C:GO:0016020 | P:long-chain fatty acid metabolic process; F:long-chain fatty acid-CoA ligase activity; F:ATP binding; C:endoplasmic reticulum; C:membrane | Long-chain-fatty-acid--CoA ligase | no GO terms | view details | ||
| JG647845.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 721 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647654.1 | Tinospora cordifolia | heat shock factor protein HSF24 | 773 | P:GO:0006357; F:GO:0000978; F:GO:0003700; C:GO:0005634 | P:regulation of transcription by RNA polymerase II; F:RNA polymerase II cis-regulatory region sequence-specific DNA binding; F:DNA-binding transcription factor activity; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding | view details | |||
| JG647851.1 | Tinospora cordifolia | ubiquitin family protein | 823 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG650690.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 806 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464; F:GO:0071949 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity; F:FAD binding | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG646169.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 703 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650414.1 | Tinospora cordifolia | probable LRR receptor-like serine/threonine-protein kinase At2g16250 | 794 | P:GO:0006468; F:GO:0004672; C:GO:0016020 | P:protein phosphorylation; F:protein kinase activity; C:membrane | Transferring phosphorus-containing groups | no GO terms | view details | ||
| MF459677.1 | Tinospora cordifolia | maturase K | 782 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG645624.1 | Tinospora cordifolia | glucose-1-phosphate adenylyltransferase large subunit 1 | 834 | P:GO:0005978; P:GO:0019252; F:GO:0005524; F:GO:0008878; C:GO:0009507 | P:glycogen biosynthetic process; P:starch biosynthetic process; F:ATP binding; F:glucose-1-phosphate adenylyltransferase activity; C:chloroplast | Glucose-1-phosphate adenylyltransferase | P:glycogen biosynthetic process; P:biosynthetic process; F:glucose-1-phosphate adenylyltransferase activity; F:nucleotidyltransferase activity | view details | ||
| JG649043.1 | Tinospora cordifolia | pathogen-related protein-like | 296 | no GO terms | view details | |||||
| JG646658.1 | Tinospora cordifolia | beta-glucosidase BoGH3B-like | 760 | P:GO:0009251; F:GO:0008422; C:GO:0005576 | P:glucan catabolic process; F:beta-glucosidase activity; C:extracellular region | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| LY749101.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1581 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| MS688304.1 | Tinospora cordifolia | caffeic acid 3-O-methyltransferase-like | 1095 | P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757 | P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG645634.1 | Tinospora cordifolia | Corytuberine synthase | 810 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649416.1 | Tinospora cordifolia | elongation factor 1-alpha 1-like | 825 | P:GO:0006414; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG647167.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 848 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| MH401132.1 | Tinospora cordifolia | maturase K | 634 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG651010.1 | Tinospora cordifolia | N-terminal acetyltransferase A complex auxiliary subunit NAA15 isoform X1 | 819 | P:GO:0017196; F:GO:0004596; C:GO:0005737 | P:N-terminal peptidyl-methionine acetylation; F:peptide alpha-N-acetyltransferase activity; C:cytoplasm | Acyltransferases | no GO terms | view details | ||
| JG647849.1 | Tinospora cordifolia | prolycopene isomerase, chloroplastic | 678 | P:GO:0016117; F:GO:0016491; F:GO:0046608; C:GO:0031969 | P:carotenoid biosynthetic process; F:oxidoreductase activity; F:carotenoid isomerase activity; C:chloroplast membrane | Cis-trans-isomerases; Oxidoreductases | P:carotenoid metabolic process | view details | ||
| JG645831.1 | Tinospora cordifolia | protein DETOXIFICATION 33-like isoform X2 | 830 | P:GO:1990961; F:GO:0015297; F:GO:0042910; C:GO:0016021 | P:xenobiotic detoxification by transmembrane export across the plasma membrane; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:membrane | view details | ||
| JG647284.1 | Tinospora cordifolia | 60S ribosomal protein L5 | 720 | P:GO:0000027; P:GO:0006412; F:GO:0003735; F:GO:0008097; C:GO:0005634; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:structural constituent of ribosome; F:5S rRNA binding; C:nucleus; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; F:5S rRNA binding; C:ribosome | view details | |||
| JG647252.1 | Tinospora cordifolia | guanine nucleotide-binding protein subunit beta-like protein A | 837 | P:GO:0001934; P:GO:0072344; F:GO:0005080; F:GO:0043022; C:GO:0005634; C:GO:0005829; C:GO:0015935 | P:positive regulation of protein phosphorylation; P:rescue of stalled ribosome; F:protein kinase C binding; F:ribosome binding; C:nucleus; C:cytosol; C:small ribosomal subunit | no IPS match | view details | |||
| JG648269.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 694 | F:GO:0016491; F:GO:0071949; C:GO:0005576 | F:oxidoreductase activity; F:FAD binding; C:extracellular region | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650557.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 835 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647362.1 | Tinospora cordifolia | non-specific phospholipase C2 | 773 | P:GO:0009395; P:GO:0042742; P:GO:0048229; P:GO:0048364; F:GO:0004629; C:GO:0005794; C:GO:0009507; C:GO:0016021 | P:phospholipid catabolic process; P:defense response to bacterium; P:gametophyte development; P:root development; F:phospholipase C activity; C:Golgi apparatus; C:chloroplast; C:integral component of membrane | Phospholipase C; Acting on ester bonds | F:hydrolase activity, acting on ester bonds | view details | ||
| JG649488.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase-like | 767 | F:GO:0046872; F:GO:0051213 | F:metal ion binding; F:dioxygenase activity | Oxidoreductases | no GO terms | view details | ||
| JG646270.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 670 | F:GO:0016491; F:GO:0071949; C:GO:0005576 | F:oxidoreductase activity; F:FAD binding; C:extracellular region | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649525.1 | Tinospora cordifolia | protein CREG1 | 231 | C:GO:0005576; C:GO:0016021 | C:extracellular region; C:integral component of membrane | no GO terms | view details | |||
| MS687591.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 990 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG645965.1 | Tinospora cordifolia | histone deacetylase 19-like | 830 | P:GO:0006325; P:GO:0006357; P:GO:0016575; F:GO:0004407; F:GO:0046872; C:GO:0005634 | P:chromatin organization; P:regulation of transcription by RNA polymerase II; P:histone deacetylation; F:histone deacetylase activity; F:metal ion binding; C:nucleus | Histone deacetylase | P:histone deacetylation; F:histone deacetylase activity | view details | ||
| JG649200.1 | Tinospora cordifolia | Eukaryotic initiation factor 4A-15 | 807 | P:GO:0002183; F:GO:0003723; F:GO:0003724; F:GO:0003743; F:GO:0005524; F:GO:0016787; C:GO:0009507 | P:cytoplasmic translational initiation; F:RNA binding; F:RNA helicase activity; F:translation initiation factor activity; F:ATP binding; F:hydrolase activity; C:chloroplast | RNA helicase | F:nucleic acid binding; F:RNA helicase activity; F:ATP binding | view details | ||
| JG648640.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 895 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| MG946986.1 | Tinospora cordifolia | maturase K | 907 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG645872.1 | Tinospora cordifolia | FAD linked oxidase | 835 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647764.1 | Tinospora cordifolia | thaumatin-like protein | 730 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG648684.1 | Tinospora cordifolia | Adenosylhomocysteinase | 786 | P:GO:0006730; P:GO:0033353; F:GO:0004013; C:GO:0005829 | P:one-carbon metabolic process; P:S-adenosylmethionine cycle; F:adenosylhomocysteinase activity; C:cytosol | Adenosylhomocysteinase | no GO terms | view details | ||
| JG649453.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 774 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648643.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 873 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650829.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 791 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG648936.1 | Tinospora cordifolia | glycoside hydrolase family 1 protein | 772 | P:GO:0005975; P:GO:0009651; P:GO:0019762; F:GO:0008422; F:GO:0102483; F:GO:0102726; C:GO:0009507 | P:carbohydrate metabolic process; P:response to salt stress; P:glucosinolate catabolic process; F:beta-glucosidase activity; F:scopolin beta-glucosidase activity; F:DIMBOA glucoside beta-D-glucosidase activity; C:chloroplast | Beta-glucosidase; 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650617.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 765 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649948.1 | Tinospora cordifolia | ubiquitin family protein | 317 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | F:protein binding | view details | |||
| JG650695.1 | Tinospora cordifolia | 14-3-3-like protein | 842 | P:GO:0007165; C:GO:0005737 | P:signal transduction; C:cytoplasm | no GO terms | view details | |||
| JG646466.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate synthase | 758 | P:GO:0006520; P:GO:0009058; F:GO:0008483; F:GO:0030170 | P:cellular amino acid metabolic process; P:biosynthetic process; F:transaminase activity; F:pyridoxal phosphate binding | Transferring nitrogenous groups | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG649411.1 | Tinospora cordifolia | basic endochitinase-like | 757 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG645647.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase family protein | 798 | P:GO:0000271; P:GO:0006024; P:GO:0006065; F:GO:0003979; F:GO:0051287; C:GO:0005634; C:GO:0005829 | P:polysaccharide biosynthetic process; P:glycosaminoglycan biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding; C:nucleus; C:cytosol | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG646132.1 | Tinospora cordifolia | NADPH-dependent aldehyde reductase-like protein, chloroplastic | 822 | F:GO:0004316 | F:3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity | Fatty-acid synthase system; 3-oxoacyl-[acyl-carrier-protein] reductase | no GO terms | view details | ||
| JG646757.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 801 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG646522.1 | Tinospora cordifolia | Receptor-like serine/threonine-protein kinase SD1-8 | 749 | P:GO:0009987; F:GO:0004672; F:GO:0005488; C:GO:0016020 | P:cellular process; F:protein kinase activity; F:binding; C:membrane | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG650667.1 | Tinospora cordifolia | protein disulfide-isomerase | 863 | P:GO:0006457; P:GO:0034976; F:GO:0003756; C:GO:0005788 | P:protein folding; P:response to endoplasmic reticulum stress; F:protein disulfide isomerase activity; C:endoplasmic reticulum lumen | Protein disulfide-isomerase | F:protein disulfide isomerase activity | view details | ||
| JG648854.1 | Tinospora cordifolia | Adenosylhomocysteinase | 855 | P:GO:0006730; P:GO:0033353; F:GO:0004013; C:GO:0005829 | P:one-carbon metabolic process; P:S-adenosylmethionine cycle; F:adenosylhomocysteinase activity; C:cytosol | Adenosylhomocysteinase | no GO terms | view details | ||
| JG647820.1 | Tinospora cordifolia | T-complex protein 1 subunit delta | 695 | P:GO:0006457; F:GO:0005524; F:GO:0016887; F:GO:0051082; C:GO:0005832 | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; F:unfolded protein binding; C:chaperonin-containing T-complex | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| MS688303.1 | Tinospora cordifolia | caffeic acid O-methyltransferase | 1101 | P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757; F:GO:0046983 | P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG649895.1 | Tinospora cordifolia | auxin-responsive protein IAA16-like | 136 | P:GO:0006355; P:GO:0009734; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:auxin-activated signaling pathway; C:nucleus | P:regulation of transcription, DNA-templated; F:protein binding; C:nucleus | view details | |||
| JG650219.1 | Tinospora cordifolia | nitrile-specifier protein 5 | 620 | P:GO:0050790; P:GO:0080028; F:GO:0030234; C:GO:0005634 | P:regulation of catalytic activity; P:nitrile biosynthetic process; F:enzyme regulator activity; C:nucleus | F:protein binding | view details | |||
| JG648118.1 | Tinospora cordifolia | monodehydroascorbate reductase | 747 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | ||
| JG648924.1 | Tinospora cordifolia | lactoylglutathione lyase GLX1-like | 703 | P:GO:0019243; F:GO:0004462; F:GO:0046872; F:GO:0051213; C:GO:0005737 | P:methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione; F:lactoylglutathione lyase activity; F:metal ion binding; F:dioxygenase activity; C:cytoplasm | Oxidoreductases; Lactoylglutathione lyase | F:lactoylglutathione lyase activity; F:metal ion binding | view details | ||
| JG648860.1 | Tinospora cordifolia | tRNA-splicing endonuclease subunit Sen2-1 isoform X1 | 721 | P:GO:0000379; P:GO:0090502; F:GO:0000213; F:GO:0003676; C:GO:0000214 | P:tRNA-type intron splice site recognition and cleavage; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:tRNA-intron endonuclease activity; F:nucleic acid binding; C:tRNA-intron endonuclease complex | Acting on ester bonds; Acting on ester bonds | P:tRNA splicing, via endonucleolytic cleavage and ligation; F:tRNA-intron endonuclease activity; F:nucleic acid binding | view details | ||
| JG646355.1 | Tinospora cordifolia | polyubiquitin isoform X1 | 825 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG647957.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 686 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG647584.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 891 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650848.1 | Tinospora cordifolia | Phosphoenolpyruvate carboxykinase, ATP-utilizing | 787 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG647555.1 | Tinospora cordifolia | elongation factor 1-alpha | 622 | P:GO:0006414; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG646378.1 | Tinospora cordifolia | probable glutathione S-transferase | 234 | P:GO:0006749; F:GO:0004364 | P:glutathione metabolic process; F:glutathione transferase activity | Glutathione transferase | no GO terms | view details | ||
| JG647064.1 | Tinospora cordifolia | 3-ketoacyl-CoA synthase 11 | 829 | P:GO:0006633; F:GO:0102756; C:GO:0016021 | P:fatty acid biosynthetic process; F:very-long-chain 3-ketoacyl-CoA synthase activity; C:integral component of membrane | Very-long-chain 3-oxoacyl-CoA synthase | P:fatty acid biosynthetic process; F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups; C:membrane | view details | ||
| JG649007.1 | Tinospora cordifolia | aldehyde dehydrogenase family 2 member B7, mitochondrial-like | 357 | F:GO:0004029 | F:aldehyde dehydrogenase (NAD+) activity | Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG648075.1 | Tinospora cordifolia | F-box protein PP2-B15-like | 810 | F:protein binding | view details | |||||
| JG648536.1 | Tinospora cordifolia | catalase isozyme 1 | 796 | P:GO:0009845; P:GO:0042542; P:GO:0042744; P:GO:0098869; F:GO:0004096; F:GO:0020037; F:GO:0046872; C:GO:0005634; C:GO:0005886; C:GO:0009514 | P:seed germination; P:response to hydrogen peroxide; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:catalase activity; F:heme binding; F:metal ion binding; C:nucleus; C:plasma membrane; C:glyoxysome | Catalase | P:response to oxidative stress; F:catalase activity; F:heme binding | view details | ||
| JG647911.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At4g13650-like | 813 | F:GO:0003824 | F:catalytic activity | F:protein binding | view details | |||
| JG649021.1 | Tinospora cordifolia | Auxin-responsive GH3 family protein | 66 | F:GO:0016881; C:GO:0005737; C:GO:0016020; C:GO:0016021 | F:acid-amino acid ligase activity; C:cytoplasm; C:membrane; C:integral component of membrane | no IPS match | view details | |||
| JG650658.1 | Tinospora cordifolia | methionine S-methyltransferase | 780 | P:GO:0032259; P:GO:1901566; F:GO:0008483; F:GO:0030170; F:GO:0030732 | P:methylation; P:organonitrogen compound biosynthetic process; F:transaminase activity; F:pyridoxal phosphate binding; F:methionine S-methyltransferase activity | Transferring nitrogenous groups; Methionine S-methyltransferase | no GO terms | view details | ||
| JG650338.1 | Tinospora cordifolia | monodehydroascorbate reductase | 804 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG646955.1 | Tinospora cordifolia | thaumatin-like protein | 614 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649662.1 | Tinospora cordifolia | nucleobase-ascorbate transporter 12 | 777 | P:GO:0055085; F:GO:0022857; C:GO:0005886; C:GO:0016021 | P:transmembrane transport; F:transmembrane transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| JG646815.1 | Tinospora cordifolia | CDP-diacylglycerol--inositol 3-phosphatidyltransferase 1-like | 751 | P:GO:0008654; F:GO:0003881; C:GO:0016021 | P:phospholipid biosynthetic process; F:CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity; C:integral component of membrane | CDP-diacylglycerol--inositol 3-phosphatidyltransferase | P:phospholipid biosynthetic process; F:phosphotransferase activity, for other substituted phosphate groups; C:membrane | view details | ||
| JG648030.1 | Tinospora cordifolia | phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplastic | 769 | P:GO:0008652; P:GO:0009073; P:GO:0009423; F:GO:0003849; C:GO:0009507 | P:cellular amino acid biosynthetic process; P:aromatic amino acid family biosynthetic process; P:chorismate biosynthetic process; F:3-deoxy-7-phosphoheptulonate synthase activity; C:chloroplast | 3-deoxy-7-phosphoheptulonate synthase | P:aromatic amino acid family biosynthetic process; F:3-deoxy-7-phosphoheptulonate synthase activity | view details | ||
| JG645586.1 | Tinospora cordifolia | ABC transporter F family member 5 | 628 | F:GO:0005524 | F:ATP binding | F:ATP binding | view details | |||
| JG648149.1 | Tinospora cordifolia | betaine aldehyde dehydrogenase 1, chloroplastic | 777 | F:GO:0008802 | F:betaine-aldehyde dehydrogenase activity | Betaine-aldehyde dehydrogenase; Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG650984.1 | Tinospora cordifolia | Berberine bridge enzyme-like 26 | 785 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG647140.1 | Tinospora cordifolia | monodehydroascorbate reductase | 776 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG646433.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 796 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG648387.1 | Tinospora cordifolia | phosphoglucomutase, cytoplasmic | 745 | P:GO:0006006; F:GO:0000287; F:GO:0004614; C:GO:0005829 | P:glucose metabolic process; F:magnesium ion binding; F:phosphoglucomutase activity; C:cytosol | Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | P:carbohydrate metabolic process; F:phosphoglucomutase activity; F:intramolecular transferase activity, phosphotransferases | view details | ||
| JG646515.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 851 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG647578.1 | Tinospora cordifolia | basic endochitinase-like | 821 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG645942.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase 1 | 815 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG648981.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 441 | no GO terms | view details | |||||
| JG649298.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 730 | no GO terms | view details | |||||
| JG648500.1 | Tinospora cordifolia | endoglucanase 12-like | 268 | P:GO:0030245; F:GO:0008810; C:GO:0016021 | P:cellulose catabolic process; F:cellulase activity; C:integral component of membrane | Cellulase | P:carbohydrate metabolic process | view details | ||
| JG647826.1 | Tinospora cordifolia | protein ARABIDILLO 1-like | 798 | F:GO:0016874 | F:ligase activity | Ligases | no GO terms | view details | ||
| JG648049.1 | Tinospora cordifolia | probable 6-phosphogluconolactonase 2 | 807 | P:GO:0005975; P:GO:0009051; F:GO:0017057; C:GO:0005737 | P:carbohydrate metabolic process; P:pentose-phosphate shunt, oxidative branch; F:6-phosphogluconolactonase activity; C:cytoplasm | 6-phosphogluconolactonase; Carboxylesterase | P:carbohydrate metabolic process; P:pentose-phosphate shunt; F:6-phosphogluconolactonase activity | view details | ||
| MN186382.1 | Tinospora cordifolia | maturase K | 584 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG649115.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 813 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG650031.1 | Tinospora cordifolia | basic endochitinase-like | 817 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646400.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase-like | 802 | F:GO:0016491; F:GO:0046872 | F:oxidoreductase activity; F:metal ion binding | Oxidoreductases | no GO terms | view details | ||
| JG647751.1 | Tinospora cordifolia | methionine gamma-lyase-like | 674 | P:GO:0019343; P:GO:0019346; F:GO:0004123; F:GO:0030170; C:GO:0005737 | P:cysteine biosynthetic process via cystathionine; P:transsulfuration; F:cystathionine gamma-lyase activity; F:pyridoxal phosphate binding; C:cytoplasm | Cystathionine gamma-lyase | P:transsulfuration; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG649062.1 | Tinospora cordifolia | basic endochitinase-like | 583 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061; C:GO:0005576; C:GO:0005773 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding; C:extracellular region; C:vacuole | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647131.1 | Tinospora cordifolia | pyruvate decarboxylase 2 | 855 | F:GO:0000287; F:GO:0004737; F:GO:0030976; C:GO:0005829 | F:magnesium ion binding; F:pyruvate decarboxylase activity; F:thiamine pyrophosphate binding; C:cytosol | Pyruvate decarboxylase | F:magnesium ion binding; F:carboxy-lyase activity; F:thiamine pyrophosphate binding | view details | ||
| JG646611.1 | Tinospora cordifolia | linamarin synthase 2-like | 686 | P:GO:0009718; F:GO:0047213; F:GO:0080043; F:GO:0080044 | P:anthocyanin-containing compound biosynthetic process; F:anthocyanidin 3-O-glucosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Anthocyanidin 3-O-glucosyltransferase | no GO terms | view details | ||
| MS687778.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1617 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646881.1 | Tinospora cordifolia | Oxalate--CoA ligase | 473 | P:GO:0006631; P:GO:0009698; F:GO:0008756; F:GO:0016207; F:GO:0031956; C:GO:0016021 | P:fatty acid metabolic process; P:phenylpropanoid metabolic process; F:o-succinylbenzoate-CoA ligase activity; F:4-coumarate-CoA ligase activity; F:medium-chain fatty acid-CoA ligase activity; C:integral component of membrane | 4-coumarate--CoA ligase; o-succinylbenzoate--CoA ligase | no GO terms | view details | ||
| JG647968.1 | Tinospora cordifolia | Elongation factor 2 | 673 | P:GO:0006414; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTP binding | view details | ||
| JG650419.1 | Tinospora cordifolia | enolase | 835 | P:GO:0006096; F:GO:0000287; F:GO:0004634; C:GO:0000015 | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | Phosphopyruvate hydratase | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | view details | ||
| JG648666.1 | Tinospora cordifolia | basic endochitinase | 784 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649184.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 796 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG647734.1 | Tinospora cordifolia | Kh domain-containing protein | 763 | P:GO:0048024; F:GO:0003729; C:GO:0005634 | P:regulation of mRNA splicing, via spliceosome; F:mRNA binding; C:nucleus | F:RNA binding | view details | |||
| JG646653.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1-like | 767 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | no GO terms | view details | ||
| LY749100.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1569 | P:GO:0006259; P:GO:0006974; P:GO:0009058; P:GO:0051276; P:GO:0098542; F:GO:0000166; F:GO:0004386; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:DNA metabolic process; P:cellular response to DNA damage stimulus; P:biosynthetic process; P:chromosome organization; P:defense response to other organism; F:nucleotide binding; F:helicase activity; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647423.1 | Tinospora cordifolia | berberine bridge enzyme-like 18 | 783 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648889.1 | Tinospora cordifolia | basic endochitinase | 791 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648929.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 734 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650320.1 | Tinospora cordifolia | polyubiquitin 3 | 662 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0016874; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:ligase activity; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | Ligases | F:protein binding | view details | ||
| JG645731.1 | Tinospora cordifolia | probable aldo-keto reductase 2 | 741 | F:GO:0016491 | F:oxidoreductase activity | Oxidoreductases | no IPS match | view details | ||
| JG649221.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 717 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645644.1 | Tinospora cordifolia | polyubiquitin isoform X1 | 825 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG648267.1 | Tinospora cordifolia | uncharacterized PKHD-type hydroxylase At1g22950-like | 488 | F:GO:0005506; F:GO:0016705; F:GO:0031418; F:GO:0051213; C:GO:0016021 | F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:L-ascorbic acid binding; F:dioxygenase activity; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no GO terms | view details | ||
| JG647281.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 558 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | ||
| JG649968.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 207 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | no GO terms | view details | ||
| JG647520.1 | Tinospora cordifolia | extradiol ring-cleavage dioxygenase | 765 | P:GO:0006725; F:GO:0008198; F:GO:0008270; F:GO:0016701; F:GO:0051213 | P:cellular aromatic compound metabolic process; F:ferrous iron binding; F:zinc ion binding; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen; F:dioxygenase activity | Acting on single donors with incorporation of molecular oxygen (oxygenases). The oxygen incorporated need not be derived from O(2) | P:cellular aromatic compound metabolic process; F:ferrous iron binding; F:zinc ion binding; F:oxidoreductase activity; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | view details | ||
| JG649725.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 658 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG646924.1 | Tinospora cordifolia | probable glutathione S-transferase | 619 | P:GO:0006749; P:GO:0009636; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; P:response to toxic substance; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG650529.1 | Tinospora cordifolia | phosphatase IMPL1, chloroplastic | 767 | P:GO:0006021; P:GO:0007165; P:GO:0046854; P:GO:0046855; F:GO:0008934; F:GO:0046872; F:GO:0052832; F:GO:0052833 | P:inositol biosynthetic process; P:signal transduction; P:phosphatidylinositol phosphate biosynthetic process; P:inositol phosphate dephosphorylation; F:inositol monophosphate 1-phosphatase activity; F:metal ion binding; F:inositol monophosphate 3-phosphatase activity; F:inositol monophosphate 4-phosphatase activity | Inositol-phosphate phosphatase | P:phosphatidylinositol phosphate biosynthetic process; P:inositol phosphate dephosphorylation | view details | ||
| JG645934.1 | Tinospora cordifolia | 26S protease regulatory subunit S10B homolog B | 749 | P:GO:0030433; P:GO:0045899; P:GO:1901800; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0036402; C:GO:0005634; C:GO:0008540; C:GO:0031597 | P:ubiquitin-dependent ERAD pathway; P:positive regulation of RNA polymerase II transcription preinitiation complex assembly; P:positive regulation of proteasomal protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:proteasome-activating activity; C:nucleus; C:proteasome regulatory particle, base subcomplex; C:cytosolic proteasome complex | Acting on acid anhydrides; Acting on peptide bonds (peptidases); Proteasome ATPase | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG646712.1 | Tinospora cordifolia | 2-isopropylmalate synthase A-like | 779 | P:GO:0009098; F:GO:0003852 | P:leucine biosynthetic process; F:2-isopropylmalate synthase activity | 2-isopropylmalate synthase | F:catalytic activity | view details | ||
| JG650751.1 | Tinospora cordifolia | polygalacturonase inhibitor-like | 808 | P:GO:0016310; F:GO:0016301; C:GO:0110165 | P:phosphorylation; F:kinase activity; C:cellular anatomical entity | Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JX125078.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 588 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG649283.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 783 | no GO terms | view details | |||||
| JG647807.1 | Tinospora cordifolia | ubiquitin family protein | 809 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG647956.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 736 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG646818.1 | Tinospora cordifolia | indole-3-acetaldehyde oxidase-like | 729 | F:iron ion binding; F:oxidoreductase activity | view details | |||||
| JG645748.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 818 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650846.1 | Tinospora cordifolia | gamma aminobutyrate transaminase 3, chloroplastic isoform X2 | 648 | P:GO:0009102; P:GO:0009448; F:GO:0003867; F:GO:0004015; F:GO:0030170 | P:biotin biosynthetic process; P:gamma-aminobutyric acid metabolic process; F:4-aminobutyrate transaminase activity; F:adenosylmethionine-8-amino-7-oxononanoate transaminase activity; F:pyridoxal phosphate binding | Adenosylmethionine--8-amino-7-oxononanoate transaminase | F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG649753.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 598 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG646910.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 543 | no GO terms | view details | |||||
| JG646704.1 | Tinospora cordifolia | U-box domain-containing protein 27 | 803 | F:GO:0004842 | F:ubiquitin-protein transferase activity | Transferases | P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:ubiquitin protein ligase activity | view details | ||
| JG648026.1 | Tinospora cordifolia | probable flavin-containing monooxygenase 1 | 828 | F:GO:0004499; F:GO:0050660; F:GO:0050661 | F:N,N-dimethylaniline monooxygenase activity; F:flavin adenine dinucleotide binding; F:NADP binding | Flavin-containing monooxygenase | F:N,N-dimethylaniline monooxygenase activity; F:flavin adenine dinucleotide binding; F:NADP binding | view details | ||
| JG649653.1 | Tinospora cordifolia | enolase | 818 | P:GO:0006096; F:GO:0000287; F:GO:0004634; C:GO:0000015 | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | Phosphopyruvate hydratase | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | view details | ||
| JG645661.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 670 | P:GO:0009058; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647290.1 | Tinospora cordifolia | cationic amino acid transporter 1-like | 667 | P:GO:0015813; P:GO:1903401; F:GO:0005313; F:GO:0015189; C:GO:0005886; C:GO:0016021 | P:L-glutamate transmembrane transport; P:L-lysine transmembrane transport; F:L-glutamate transmembrane transporter activity; F:L-lysine transmembrane transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | no GO terms | view details | ||
| JG645753.1 | Tinospora cordifolia | 2-oxoglutarate-dependent dioxygenase DAO-like | 739 | P:GO:0009852; P:GO:0010252; P:GO:1901576; F:GO:0016705; F:GO:0046872; F:GO:0050302; F:GO:0051213 | P:auxin catabolic process; P:auxin homeostasis; P:organic substance biosynthetic process; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:metal ion binding; F:indole-3-acetaldehyde oxidase activity; F:dioxygenase activity | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Aryl-aldehyde oxidase; Indole-3-acetaldehyde oxidase; Aldehyde oxidase | no GO terms | view details | ||
| JG651001.1 | Tinospora cordifolia | E3 ubiquitin-protein ligase UPL2-like isoform X1 | 845 | P:GO:0000209; P:GO:0006414; P:GO:0043161; P:GO:0045732; F:GO:0003746; F:GO:0061630; C:GO:0005737 | P:protein polyubiquitination; P:translational elongation; P:proteasome-mediated ubiquitin-dependent protein catabolic process; P:positive regulation of protein catabolic process; F:translation elongation factor activity; F:ubiquitin protein ligase activity; C:cytoplasm | Transferases | no GO terms | view details | ||
| JG649384.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 778 | P:GO:0005975; F:GO:0004497; F:GO:0004553; F:GO:0016705; F:GO:0016765; F:GO:0046872; C:GO:0046658 | P:carbohydrate metabolic process; F:monooxygenase activity; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:metal ion binding; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646798.1 | Tinospora cordifolia | Corytuberine synthase | 848 | P:GO:0019438; P:GO:0071704; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:aromatic compound biosynthetic process; P:organic substance metabolic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646534.1 | Tinospora cordifolia | 14-3-3-like protein | 776 | P:GO:0007165; C:GO:0005737 | P:signal transduction; C:cytoplasm | no GO terms | view details | |||
| JG647974.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 810 | P:GO:0006508; F:GO:0004190; C:GO:0016021 | P:proteolysis; F:aspartic-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG649070.1 | Tinospora cordifolia | actin-like | 722 | F:GO:0005524; C:GO:0005737; C:GO:0005856 | F:ATP binding; C:cytoplasm; C:cytoskeleton | no GO terms | view details | |||
| JG649510.1 | Tinospora cordifolia | prostaglandin reductase-3 | 813 | P:GO:0006979; F:GO:0008270; F:GO:0016491 | P:response to oxidative stress; F:zinc ion binding; F:oxidoreductase activity | Oxidoreductases | no GO terms | view details | ||
| JG649082.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 772 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647413.1 | Tinospora cordifolia | basic endochitinase-like | 801 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649541.1 | Tinospora cordifolia | transmembrane 9 superfamily member 3 | 747 | P:GO:0006817; P:GO:0050896; P:GO:0072657; C:GO:0000139; C:GO:0005802; C:GO:0010008; C:GO:0016021 | P:phosphate ion transport; P:response to stimulus; P:protein localization to membrane; C:Golgi membrane; C:trans-Golgi network; C:endosome membrane; C:integral component of membrane | C:integral component of membrane | view details | |||
| JG646178.1 | Tinospora cordifolia | protein MEI2-like 4 isoform X1 | 589 | F:GO:0003676; F:GO:0003723 | F:nucleic acid binding; F:RNA binding | no GO terms | view details | |||
| JG647796.1 | Tinospora cordifolia | digalactosyldiacylglycerol synthase 1, chloroplastic | 642 | P:GO:0019375; F:GO:0046481; C:GO:0009707 | P:galactolipid biosynthetic process; F:digalactosyldiacylglycerol synthase activity; C:chloroplast outer membrane | Digalactosyldiacylglycerol synthase | F:digalactosyldiacylglycerol synthase activity | view details | ||
| JG646682.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 731 | P:GO:0005975; F:GO:0004497; F:GO:0016705; F:GO:0016765; F:GO:0042973; F:GO:0046872; C:GO:0046658 | P:carbohydrate metabolic process; F:monooxygenase activity; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; F:metal ion binding; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646613.1 | Tinospora cordifolia | exocyst complex component EXO70A1-like | 770 | P:GO:0006887; P:GO:0015031; C:GO:0000145 | P:exocytosis; P:protein transport; C:exocyst | P:exocytosis; C:exocyst | view details | |||
| JG648306.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP) | 839 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG647756.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 765 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG646157.1 | Tinospora cordifolia | short-chain dehydrogenase reductase 2a | 787 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | no GO terms | view details | ||
| JG645821.1 | Tinospora cordifolia | probable acyl-activating enzyme 17, peroxisomal isoform X1 | 851 | P:GO:0009698; F:GO:0016207; C:GO:0005737; C:GO:0016021 | P:phenylpropanoid metabolic process; F:4-coumarate-CoA ligase activity; C:cytoplasm; C:integral component of membrane | 4-coumarate--CoA ligase | no GO terms | view details | ||
| JG645562.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 690 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG645805.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 867 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647324.1 | Tinospora cordifolia | basic endochitinase | 738 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650822.1 | Tinospora cordifolia | plastidic ATP/ADP-transporter-like | 787 | P:GO:0006817; P:GO:0015866; P:GO:0015867; P:GO:0050896; F:GO:0005471; F:GO:0005524; C:GO:0016021; C:GO:0031969 | P:phosphate ion transport; P:ADP transport; P:ATP transport; P:response to stimulus; F:ATP:ADP antiporter activity; F:ATP binding; C:integral component of membrane; C:chloroplast membrane | Translocases | P:nucleotide transport; F:ATP:ADP antiporter activity; C:integral component of membrane | view details | ||
| JG646941.1 | Tinospora cordifolia | wall-associated receptor kinase-like 14 | 120 | P:GO:0006468; P:GO:0007166; F:GO:0004672; F:GO:0005524; C:GO:0005886; C:GO:0016021 | P:protein phosphorylation; P:cell surface receptor signaling pathway; F:protein kinase activity; F:ATP binding; C:plasma membrane; C:integral component of membrane | Transferring phosphorus-containing groups | no IPS match | view details | ||
| JG649682.1 | Tinospora cordifolia | protein SINE1-like isoform X1 | 767 | P:GO:0000387; C:GO:0005634; C:GO:0016021; C:GO:0032797 | P:spliceosomal snRNP assembly; C:nucleus; C:integral component of membrane; C:SMN complex | no GO terms | view details | |||
| JG648153.1 | Tinospora cordifolia | berberine bridge enzyme-like 18 | 785 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG645622.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 257 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG646975.1 | Tinospora cordifolia | phenylalanine ammonia-lyase | 328 | P:GO:0006559; P:GO:0009800; F:GO:0045548; C:GO:0005737 | P:L-phenylalanine catabolic process; P:cinnamic acid biosynthetic process; F:phenylalanine ammonia-lyase activity; C:cytoplasm | Phenylalanine ammonia-lyase; Phenylalanine/tyrosine ammonia-lyase | F:catalytic activity | view details | ||
| JG646158.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 799 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647497.1 | Tinospora cordifolia | phosphatidylinositol/phosphatidylcholine transfer protein SFH8 isoform X1 | 100 | C:GO:0016021 | C:integral component of membrane | no IPS match | view details | |||
| JG649811.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 866 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG649666.1 | Tinospora cordifolia | serine/threonine-protein kinase CTR1 isoform X2 | 767 | P:GO:0001666; P:GO:0009686; P:GO:0009723; P:GO:0009744; P:GO:0009750; P:GO:0018108; P:GO:0048510; P:GO:2000035; P:GO:2000069; F:GO:0004674; F:GO:0004713; F:GO:0005524; C:GO:0005789 | P:response to hypoxia; P:gibberellin biosynthetic process; P:response to ethylene; P:response to sucrose; P:response to fructose; P:peptidyl-tyrosine phosphorylation; P:regulation of timing of transition from vegetative to reproductive phase; P:regulation of stem cell division; P:regulation of post-embryonic root development; F:protein serine/threonine kinase activity; F:protein tyrosine kinase activity; F:ATP binding; C:endoplasmic reticulum membrane | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG646671.1 | Tinospora cordifolia | sucrose nonfermenting 4-like protein | 786 | P:GO:0000266; P:GO:0009859; P:GO:0016559; P:GO:0042149; P:GO:0045859; P:GO:2000377; F:GO:0016208; F:GO:0016301; F:GO:0019887; F:GO:0019901; F:GO:1901982; C:GO:0005634; C:GO:0009569; C:GO:0031588 | P:mitochondrial fission; P:pollen hydration; P:peroxisome fission; P:cellular response to glucose starvation; P:regulation of protein kinase activity; P:regulation of reactive oxygen species metabolic process; F:AMP binding; F:kinase activity; F:protein kinase regulator activity; F:protein kinase binding; F:maltose binding; C:nucleus; C:chloroplast starch grain; C:nucleotide-activated protein kinase complex | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG646389.1 | Tinospora cordifolia | glyceraldehyde-3-phosphate dehydrogenase, cytosolic | 796 | P:GO:0006006; P:GO:0006096; F:GO:0004365; F:GO:0050661; F:GO:0051287; C:GO:0005829; C:GO:0009536 | P:glucose metabolic process; P:glycolytic process; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity; F:NADP binding; F:NAD binding; C:cytosol; C:plastid | Glyceraldehyde-3-phosphate dehydrogenase (NAD(P)(+)) (phosphorylating); Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG647399.1 | Tinospora cordifolia | cytochrome p450 82c4 | 770 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647765.1 | Tinospora cordifolia | peroxisomal adenine nucleotide carrier 1 | 322 | P:GO:0006635; P:GO:0007031; P:GO:0015866; P:GO:0015867; P:GO:0055085; F:GO:0005347; F:GO:0015217; C:GO:0005779 | P:fatty acid beta-oxidation; P:peroxisome organization; P:ADP transport; P:ATP transport; P:transmembrane transport; F:ATP transmembrane transporter activity; F:ADP transmembrane transporter activity; C:integral component of peroxisomal membrane | Translocases | P:fatty acid beta-oxidation; F:ATP transmembrane transporter activity; F:ADP transmembrane transporter activity | view details | ||
| JG647000.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 770 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648424.1 | Tinospora cordifolia | 3-ketoacyl-CoA thiolase 2, peroxisomal | 764 | P:GO:0006635; P:GO:0010124; F:GO:0003988; C:GO:0005777 | P:fatty acid beta-oxidation; P:phenylacetate catabolic process; F:acetyl-CoA C-acyltransferase activity; C:peroxisome | Acetyl-CoA C-acyltransferase | F:acyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG648779.1 | Tinospora cordifolia | basic endochitinase-like | 549 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| MH765688.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 591 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG645815.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 860 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG648580.1 | Tinospora cordifolia | MACPF domain-containing protein At4g24290-like | 743 | P:GO:0009626; P:GO:2000031 | P:plant-type hypersensitive response; P:regulation of salicylic acid mediated signaling pathway | P:defense response; P:programmed cell death; P:regulation of salicylic acid mediated signaling pathway | view details | |||
| JG647778.1 | Tinospora cordifolia | calcium-transporting ATPase 8, plasma membrane-type isoform X1 | 796 | P:GO:0070588; F:GO:0005388; F:GO:0005516; F:GO:0005524; F:GO:0016887; C:GO:0016021 | P:calcium ion transmembrane transport; F:P-type calcium transporter activity; F:calmodulin binding; F:ATP binding; F:ATP hydrolysis activity; C:integral component of membrane | Acting on acid anhydrides; P-type Ca(2+) transporter | F:nucleotide binding; F:transporter activity; F:ATP binding; F:ATP hydrolysis activity; C:integral component of membrane | view details | ||
| JG649719.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 781 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650917.1 | Tinospora cordifolia | Aldehyde dehydrogenase family 7 member a1 | 719 | F:GO:0004029; F:GO:0043878 | F:aldehyde dehydrogenase (NAD+) activity; F:glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity | Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:aldehyde dehydrogenase (NAD+) activity; F:oxidoreductase activity | view details | ||
| JG647219.1 | Tinospora cordifolia | cell division cycle protein 48 homolog | 697 | P:GO:0030433; P:GO:0030970; P:GO:0051228; P:GO:0051301; P:GO:0071712; P:GO:0097352; F:GO:0005524; F:GO:0008237; F:GO:0016887; F:GO:0031593; C:GO:0005634; C:GO:0005829; C:GO:0016021; C:GO:0034098 | P:ubiquitin-dependent ERAD pathway; P:retrograde protein transport, ER to cytosol; P:mitotic spindle disassembly; P:cell division; P:ER-associated misfolded protein catabolic process; P:autophagosome maturation; F:ATP binding; F:metallopeptidase activity; F:ATP hydrolysis activity; F:polyubiquitin modification-dependent protein binding; C:nucleus; C:cytosol; C:integral component of membrane; C:VCP-NPL4-UFD1 AAA ATPase complex | Acting on acid anhydrides; Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| JG648272.1 | Tinospora cordifolia | basic endochitinase-like | 110 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | no IPS match | view details | ||
| MS688352.1 | Tinospora cordifolia | (+)-neomenthol dehydrogenase-like | 927 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG648883.1 | Tinospora cordifolia | probable cytosolic oligopeptidase A | 644 | P:GO:0006508; P:GO:0006518; F:GO:0004222; F:GO:0046872; C:GO:0016021 | P:proteolysis; P:peptide metabolic process; F:metalloendopeptidase activity; F:metal ion binding; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:metalloendopeptidase activity | view details | ||
| MA258229.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1569 | P:GO:0006259; P:GO:0006974; P:GO:0009058; P:GO:0051276; P:GO:0098542; F:GO:0000166; F:GO:0004386; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:DNA metabolic process; P:cellular response to DNA damage stimulus; P:biosynthetic process; P:chromosome organization; P:defense response to other organism; F:nucleotide binding; F:helicase activity; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649945.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 505 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG647358.1 | Tinospora cordifolia | probable pectinesterase/pectinesterase inhibitor 7 | 824 | F:GO:0030599; F:GO:0046910; C:GO:0016020 | F:pectinesterase activity; F:pectinesterase inhibitor activity; C:membrane | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG645694.1 | Tinospora cordifolia | EP1-like glycoprotein 2 | 847 | F:GO:0016829; F:GO:0030246; C:GO:0110165 | F:lyase activity; F:carbohydrate binding; C:cellular anatomical entity | Lyases | no GO terms | view details | ||
| JG650205.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1-like | 772 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | no GO terms | view details | ||
| JG645558.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 765 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648855.1 | Tinospora cordifolia | peptidyl-prolyl cis-trans isomerase-like | 199 | P:GO:0000413; P:GO:0006457; F:GO:0003755; C:GO:0110165 | P:protein peptidyl-prolyl isomerization; P:protein folding; F:peptidyl-prolyl cis-trans isomerase activity; C:cellular anatomical entity | Peptidylprolyl isomerase | P:protein peptidyl-prolyl isomerization; F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG646893.1 | Tinospora cordifolia | pathogen-related protein-like | 484 | no GO terms | view details | |||||
| JG646978.1 | Tinospora cordifolia | basic endochitinase-like | 697 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | no GO terms | view details | ||
| JG646332.1 | Tinospora cordifolia | serine/threonine-protein kinase AtPK2/AtPK19-like | 610 | P:GO:0018105; F:GO:0004674; F:GO:0005524; C:GO:0005634; C:GO:0005737 | P:peptidyl-serine phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:protein serine/threonine kinase activity; F:ATP binding | view details | ||
| JG647322.1 | Tinospora cordifolia | ruBisCO large subunit-binding protein subunit beta, chloroplastic | 647 | P:GO:0042026; F:GO:0005524; F:GO:0016887; C:GO:0005737 | P:protein refolding; F:ATP binding; F:ATP hydrolysis activity; C:cytoplasm | Acting on acid anhydrides | no IPS match | view details | ||
| JG648990.1 | Tinospora cordifolia | copper-transporting ATPase RAN1 | 207 | P:GO:0035434; P:GO:0055070; F:GO:0005507; F:GO:0005524; F:GO:0016887; F:GO:0043682; C:GO:0016021 | P:copper ion transmembrane transport; P:copper ion homeostasis; F:copper ion binding; F:ATP binding; F:ATP hydrolysis activity; F:P-type divalent copper transporter activity; C:integral component of membrane | Acting on acid anhydrides; P-type Cu(2+) transporter | no GO terms | view details | ||
| JG645741.1 | Tinospora cordifolia | malonyl-coenzyme A:anthocyanin 3-O-glucoside-6'-O-malonyltransferase-like | 839 | F:GO:0016740; F:GO:0016747 | F:transferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | no GO terms | view details | |||
| JG648482.1 | Tinospora cordifolia | probable L-gulonolactone oxidase 6 | 788 | P:GO:0019853; F:GO:0003885; F:GO:0050105; F:GO:0071949; C:GO:0016020 | P:L-ascorbic acid biosynthetic process; F:D-arabinono-1,4-lactone oxidase activity; F:L-gulonolactone oxidase activity; F:FAD binding; C:membrane | D-arabinono-1,4-lactone oxidase; L-gulonolactone oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649338.1 | Tinospora cordifolia | protein QUIRKY | 824 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650823.1 | Tinospora cordifolia | SNF1-related protein kinase catalytic subunit alpha KIN10 | 857 | P:GO:0006468; P:GO:0035556; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310 | P:protein phosphorylation; P:intracellular signal transduction; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity | Dual-specificity kinase | P:protein phosphorylation; F:protein kinase activity; F:protein binding; F:ATP binding | view details | ||
| JG648584.1 | Tinospora cordifolia | probable 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 642 | P:GO:0016126; F:GO:0000247; F:GO:0004769; F:GO:0047750; C:GO:0005789; C:GO:0016021 | P:sterol biosynthetic process; F:C-8 sterol isomerase activity; F:steroid delta-isomerase activity; F:cholestenol delta-isomerase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | Steroid Delta-isomerase; Cholestenol Delta-isomerase | P:sterol metabolic process; F:cholestenol delta-isomerase activity; C:integral component of membrane | view details | ||
| JG648044.1 | Tinospora cordifolia | inactive beta-amylase 9 | 802 | P:GO:0000272; F:GO:0016161; F:GO:0102229 | P:polysaccharide catabolic process; F:beta-amylase activity; F:amylopectin maltohydrolase activity | Beta-amylase | P:polysaccharide catabolic process; F:beta-amylase activity | view details | ||
| JG647507.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 768 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648910.1 | Tinospora cordifolia | Berberine bridge enzyme-like 26 | 767 | F:GO:0050328; F:GO:0071949; C:GO:0016021 | F:tetrahydroberberine oxidase activity; F:FAD binding; C:integral component of membrane | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG645877.1 | Tinospora cordifolia | 2-oxoisovalerate dehydrogenase subunit alpha 2, mitochondrial | 813 | P:GO:0009083; F:GO:0016624; C:GO:0005947 | P:branched-chain amino acid catabolic process; F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor; C:mitochondrial alpha-ketoglutarate dehydrogenase complex | Acting on the aldehyde or oxo group of donors | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor | view details | ||
| JG647533.1 | Tinospora cordifolia | puromycin-sensitive aminopeptidase isoform X3 | 649 | P:GO:0006508; F:GO:0004177; F:GO:0008237; F:GO:0008270 | P:proteolysis; F:aminopeptidase activity; F:metallopeptidase activity; F:zinc ion binding | Acting on peptide bonds (peptidases) | P:proteolysis; F:metallopeptidase activity; F:zinc ion binding | view details | ||
| JG647319.1 | Tinospora cordifolia | basic endochitinase-like | 858 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646934.1 | Tinospora cordifolia | BES1/BZR1 homolog protein 2-like | 210 | P:GO:0006355; P:GO:0009742; F:GO:0003700; C:GO:0005634; C:GO:0005737 | P:regulation of transcription, DNA-templated; P:brassinosteroid mediated signaling pathway; F:DNA-binding transcription factor activity; C:nucleus; C:cytoplasm | no IPS match | view details | |||
| JG650674.1 | Tinospora cordifolia | probable ascorbate-specific transmembrane electron transporter 1 | 777 | F:GO:0016491; C:GO:0016021 | F:oxidoreductase activity; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity | view details | ||
| JG650959.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 737 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG650185.1 | Tinospora cordifolia | Berberine bridge enzyme-like 26 | 749 | F:GO:0050328; F:GO:0071949; C:GO:0016021 | F:tetrahydroberberine oxidase activity; F:FAD binding; C:integral component of membrane | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646572.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 779 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650124.1 | Tinospora cordifolia | malate dehydrogenase | 758 | P:GO:0006099; P:GO:0006107; P:GO:0006108; P:GO:0006734; F:GO:0030060 | P:tricarboxylic acid cycle; P:oxaloacetate metabolic process; P:malate metabolic process; P:NADH metabolic process; F:L-malate dehydrogenase activity | Malate dehydrogenase | P:malate metabolic process; F:catalytic activity; F:oxidoreductase activity; F:malate dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG647996.1 | Tinospora cordifolia | SKP1-like protein 1B | 503 | P:GO:0016310; P:GO:0016567; P:GO:0031146; F:GO:0016301; F:GO:0097602; C:GO:0005634; C:GO:0005737 | P:phosphorylation; P:protein ubiquitination; P:SCF-dependent proteasomal ubiquitin-dependent protein catabolic process; F:kinase activity; F:cullin family protein binding; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | P:ubiquitin-dependent protein catabolic process | view details | ||
| JG646915.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 505 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG647436.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP) | 773 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| MK244676.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 481 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG648650.1 | Tinospora cordifolia | basic endochitinase-like | 821 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647469.1 | Tinospora cordifolia | methyltransferase-like protein 2 isoform X2 | 712 | P:GO:0032259; F:GO:0008168; C:GO:0005634 | P:methylation; F:methyltransferase activity; C:nucleus | Transferring one-carbon groups | no GO terms | view details | ||
| MS688297.1 | Tinospora cordifolia | (RS)-norcoclaurine 6-O-methyltransferase | 1056 | P:GO:0009821; P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0030786; F:GO:0042802; F:GO:0046983 | P:alkaloid biosynthetic process; P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:(RS)-norcoclaurine 6-O-methyltransferase activity; F:identical protein binding; F:protein dimerization activity | (RS)-norcoclaurine 6-O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG648131.1 | Tinospora cordifolia | chitinase precursor | 134 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity | Chitinase | no IPS match | view details | ||
| JG647091.1 | Tinospora cordifolia | protein GRIP | 741 | P:GO:0090502; F:GO:0003676; F:GO:0004523 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:nucleic acid binding; F:RNA-DNA hybrid ribonuclease activity | Acting on ester bonds; Ribonuclease H | no GO terms | view details | ||
| JG650139.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | 672 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; C:chloroplast | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG650102.1 | Tinospora cordifolia | FAD linked oxidase | 763 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG646694.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 791 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG648921.1 | Tinospora cordifolia | basic endochitinase | 778 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648776.1 | Tinospora cordifolia | porphobilinogen deaminase, chloroplastic | 455 | P:GO:0006782; P:GO:0018160; F:GO:0004418; C:GO:0005737; C:GO:0016021 | P:protoporphyrinogen IX biosynthetic process; P:peptidyl-pyrromethane cofactor linkage; F:hydroxymethylbilane synthase activity; C:cytoplasm; C:integral component of membrane | Hydroxymethylbilane synthase | P:tetrapyrrole biosynthetic process; F:hydroxymethylbilane synthase activity | view details | ||
| JG645552.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 720 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| MH765687.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 591 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG650784.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 866 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464; F:GO:0071949 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity; F:FAD binding | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| MS687901.1 | Tinospora cordifolia | S-norcoclaurine synthase 1-like | 1083 | F:GO:0016491; F:GO:0016829 | F:oxidoreductase activity; F:lyase activity | Oxidoreductases; Lyases | no GO terms | view details | ||
| JG648807.1 | Tinospora cordifolia | probable aldo-keto reductase 2 | 846 | F:GO:0004033; C:GO:0005737 | F:aldo-keto reductase (NADP) activity; C:cytoplasm | Acting on the CH-OH group of donors | F:oxidoreductase activity | view details | ||
| JG650919.1 | Tinospora cordifolia | berberine bridge enzyme | 703 | P:GO:0009820; F:GO:0050328; F:GO:0050468; F:GO:0071949; C:GO:0031410 | P:alkaloid metabolic process; F:tetrahydroberberine oxidase activity; F:reticuline oxidase activity; F:FAD binding; C:cytoplasmic vesicle | Reticuline oxidase; Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG645832.1 | Tinospora cordifolia | putative serine/threonine-protein kinase-like protein CCR3 | 815 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:protein serine/threonine kinase activity; F:ATP binding | view details | ||
| JG646227.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 767 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649098.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 683 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG648896.1 | Tinospora cordifolia | acyl-coenzyme A oxidase 4, peroxisomal | 664 | P:GO:0006635; F:GO:0003995; F:GO:0050660; C:GO:0005777; C:GO:0016021 | P:fatty acid beta-oxidation; F:acyl-CoA dehydrogenase activity; F:flavin adenine dinucleotide binding; C:peroxisome; C:integral component of membrane | Acting on the CH-CH group of donors | P:fatty acid beta-oxidation; F:acyl-CoA dehydrogenase activity; F:oxidoreductase activity, acting on the CH-CH group of donors; F:flavin adenine dinucleotide binding | view details | ||
| JG646042.1 | Tinospora cordifolia | Detoxification-like protein | 719 | P:GO:1990961; F:GO:0015297; F:GO:0042910; C:GO:0016021 | P:xenobiotic detoxification by transmembrane export across the plasma membrane; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:membrane | view details | ||
| JG648965.1 | Tinospora cordifolia | probable L-type lectin-domain containing receptor kinase I.6 | 211 | P:GO:0002229; P:GO:0016310; P:GO:0042742; P:GO:0050794; F:GO:0000166; F:GO:0004675; C:GO:0005886 | P:defense response to oomycetes; P:phosphorylation; P:defense response to bacterium; P:regulation of cellular process; F:nucleotide binding; F:transmembrane receptor protein serine/threonine kinase activity; C:plasma membrane | Receptor protein serine/threonine kinase | F:carbohydrate binding | view details | ||
| JG645758.1 | Tinospora cordifolia | basic endochitinase-like | 817 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646422.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 117 | P:GO:0009416; F:GO:0016881; C:GO:0005737; C:GO:0016021 | P:response to light stimulus; F:acid-amino acid ligase activity; C:cytoplasm; C:integral component of membrane | Forming carbon-nitrogen bonds | no IPS match | view details | ||
| JG647118.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 828 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG647313.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 728 | P:GO:0009058; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648117.1 | Tinospora cordifolia | UPF0496 protein 4-like | 847 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG650050.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 766 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| MS688302.1 | Tinospora cordifolia | caffeic acid 3-O-methyltransferase | 1098 | P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0008757; F:GO:0046983 | P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; F:protein dimerization activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG647382.1 | Tinospora cordifolia | Sulfate transporter 3.1 | 602 | P:GO:1902358; F:GO:0008271; F:GO:0015301; C:GO:0005887; C:GO:0009507 | P:sulfate transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane; C:chloroplast | Translocases | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| MG946985.1 | Tinospora cordifolia | maturase K | 857 | P:GO:0006397; P:GO:0008033; P:GO:0008380; F:GO:0003723; C:GO:0009507 | P:mRNA processing; P:tRNA processing; P:RNA splicing; F:RNA binding; C:chloroplast | P:mRNA processing; C:chloroplast | view details | |||
| JG650265.1 | Tinospora cordifolia | protein DETOXIFICATION 12-like | 832 | P:GO:1990961; F:GO:0015297; F:GO:0042910; C:GO:0016021 | P:xenobiotic detoxification by transmembrane export across the plasma membrane; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; P:xenobiotic detoxification by transmembrane export across the plasma membrane; F:antiporter activity; F:xenobiotic transmembrane transporter activity; C:membrane | view details | ||
| JG645522.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 821 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG649833.1 | Tinospora cordifolia | Indole-3-acetic acid-amido synthetase GH3.6 | 838 | F:GO:0010279; C:GO:0005737 | F:indole-3-acetic acid amido synthetase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650692.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 841 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG646133.1 | Tinospora cordifolia | chaperone protein ClpD, chloroplastic | 795 | F:GO:0005524; F:GO:0016887; C:GO:0009570 | F:ATP binding; F:ATP hydrolysis activity; C:chloroplast stroma | Acting on acid anhydrides | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG650434.1 | Tinospora cordifolia | BURP domain protein RD22-like | 773 | no GO terms | view details | |||||
| JG649031.1 | Tinospora cordifolia | protein MAIN-LIKE 1-like | 235 | P:GO:0010073; P:GO:0048507 | P:meristem maintenance; P:meristem development | no IPS match | view details | |||
| JG650938.1 | Tinospora cordifolia | Corytuberine synthase | 819 | P:GO:0009058; P:GO:0071704; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:biosynthetic process; P:organic substance metabolic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647039.1 | Tinospora cordifolia | polyphenol oxidase | 736 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:catechol oxidase activity; F:oxidoreductase activity | view details | ||
| JG649941.1 | Tinospora cordifolia | aspartate aminotransferase, cytoplasmic | 423 | P:GO:0006520; P:GO:0010150; P:GO:1901566; F:GO:0004069; F:GO:0030170; C:GO:0005739; C:GO:0005777; C:GO:0009536 | P:cellular amino acid metabolic process; P:leaf senescence; P:organonitrogen compound biosynthetic process; F:L-aspartate:2-oxoglutarate aminotransferase activity; F:pyridoxal phosphate binding; C:mitochondrion; C:peroxisome; C:plastid | Aspartate transaminase | P:cellular amino acid metabolic process; P:biosynthetic process; F:catalytic activity; F:transaminase activity; F:pyridoxal phosphate binding | view details | ||
| JG648517.1 | Tinospora cordifolia | basic endochitinase | 789 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646106.1 | Tinospora cordifolia | Alpha/beta hydrolase fold-1 | 772 | F:GO:0016787 | F:hydrolase activity | Hydrolases | F:catalytic activity | view details | ||
| JG648307.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 822 | P:GO:0009733; F:GO:0016881; C:GO:0005737 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| MS687590.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 960 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG647369.1 | Tinospora cordifolia | respiratory burst oxidase homolog protein D | 763 | P:GO:0098869; F:GO:0000293; F:GO:0004601; F:GO:0005509; F:GO:0016174; C:GO:0005886; C:GO:0016021 | P:cellular oxidant detoxification; F:ferric-chelate reductase activity; F:peroxidase activity; F:calcium ion binding; F:NAD(P)H oxidase H2O2-forming activity; C:plasma membrane; C:integral component of membrane | NAD(P)H oxidase (H(2)O(2)-forming); Oxidizing metal ions; Acting on a peroxide as acceptor | F:oxidoreductase activity; C:membrane | view details | ||
| JG649228.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 714 | P:GO:0005975; F:GO:0004497; F:GO:0004553; F:GO:0016705; F:GO:0016765; F:GO:0046872; C:GO:0046658 | P:carbohydrate metabolic process; F:monooxygenase activity; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:metal ion binding; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648512.1 | Tinospora cordifolia | glutamine synthetase nodule isozyme-like | 708 | P:GO:0006542; F:GO:0004356; F:GO:0005524; C:GO:0005737 | P:glutamine biosynthetic process; F:glutamate-ammonia ligase activity; F:ATP binding; C:cytoplasm | Glutamine synthetase | P:glutamine biosynthetic process; P:nitrogen compound metabolic process; F:catalytic activity; F:glutamate-ammonia ligase activity | view details | ||
| JG649287.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 764 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| MT834944.1 | Tinospora cordifolia | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 1449 | P:GO:0009853; P:GO:0019253; F:GO:0000287; F:GO:0004497; F:GO:0016984; C:GO:0009507 | P:photorespiration; P:reductive pentose-phosphate cycle; F:magnesium ion binding; F:monooxygenase activity; F:ribulose-bisphosphate carboxylase activity; C:chloroplast | Ribulose-bisphosphate carboxylase; Oxidoreductases | P:carbon fixation; F:magnesium ion binding; F:ribulose-bisphosphate carboxylase activity | view details | ||
| JG646177.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 451 | P:GO:0005975; F:GO:0016765; F:GO:0042973 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG648622.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 742 | F:GO:0004032; F:GO:0016746; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; F:acyltransferase activity; C:cytosol | Aldose reductase; Acyltransferases | F:oxidoreductase activity | view details | ||
| JG647315.1 | Tinospora cordifolia | pyruvate dehydrogenase E1 component subunit alpha, mitochondrial | 780 | P:GO:0006086; P:GO:0006096; F:GO:0004739; C:GO:0005737; C:GO:0043231 | P:acetyl-CoA biosynthetic process from pyruvate; P:glycolytic process; F:pyruvate dehydrogenase (acetyl-transferring) activity; C:cytoplasm; C:intracellular membrane-bounded organelle | Pyruvate dehydrogenase (acetyl-transferring) | F:oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor | view details | ||
| JG647950.1 | Tinospora cordifolia | basic endochitinase-like | 767 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG648897.1 | Tinospora cordifolia | L-ascorbate peroxidase, cytosolic | 751 | P:GO:0009723; P:GO:0034599; P:GO:0042542; P:GO:0042744; P:GO:0045454; P:GO:0090378; P:GO:0098869; F:GO:0016688; F:GO:0020037; F:GO:0046872; C:GO:0009507 | P:response to ethylene; P:cellular response to oxidative stress; P:response to hydrogen peroxide; P:hydrogen peroxide catabolic process; P:cell redox homeostasis; P:seed trichome elongation; P:cellular oxidant detoxification; F:L-ascorbate peroxidase activity; F:heme binding; F:metal ion binding; C:chloroplast | L-ascorbate peroxidase | P:response to oxidative stress; P:cellular response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG649121.1 | Tinospora cordifolia | basic endochitinase-like | 635 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649129.1 | Tinospora cordifolia | FAD linked oxidase | 844 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650271.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 817 | P:GO:0009733; F:GO:0016881; C:GO:0005737; C:GO:0016021 | P:response to auxin; F:acid-amino acid ligase activity; C:cytoplasm; C:integral component of membrane | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649902.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At1g74750-like | 109 | no IPS match | view details | |||||
| JG646220.1 | Tinospora cordifolia | far upstream element-binding protein 1-like | 743 | P:GO:0010468; F:GO:0003729; F:GO:0016740; C:GO:0005634; C:GO:0005737 | P:regulation of gene expression; F:mRNA binding; F:transferase activity; C:nucleus; C:cytoplasm | Transferases | F:RNA binding | view details | ||
| JG649937.1 | Tinospora cordifolia | osmotin-like protein osm34 | 342 | P:GO:0006952; C:GO:0016020 | P:defense response; C:membrane | no GO terms | view details | |||
| JG650025.1 | Tinospora cordifolia | Nadp-dependent glyceraldehyde-3-phosphate dehydrogenase | 671 | F:GO:0008911 | F:lactaldehyde dehydrogenase activity | Lactaldehyde dehydrogenase; Aldehyde dehydrogenase (NAD(P)(+)); Aldehyde dehydrogenase (NAD(+)) | F:oxidoreductase activity | view details | ||
| JG647979.1 | Tinospora cordifolia | glycerophosphodiester phosphodiesterase GDPDL3-like | 691 | P:GO:0006071; P:GO:0006629; F:GO:0008889 | P:glycerol metabolic process; P:lipid metabolic process; F:glycerophosphodiester phosphodiesterase activity | Glycerophosphodiester phosphodiesterase | P:lipid metabolic process; F:phosphoric diester hydrolase activity | view details | ||
| JG648377.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 809 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG647164.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 790 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648412.1 | Tinospora cordifolia | caffeoylshikimate esterase-like | 794 | F:GO:0016298; C:GO:0016020 | F:lipase activity; C:membrane | Acting on ester bonds | no GO terms | view details | ||
| JG650740.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 777 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG649382.1 | Tinospora cordifolia | COBRA-like protein 7 | 725 | P:GO:0010215; F:GO:0030246; C:GO:0016021; C:GO:0031225 | P:cellulose microfibril organization; F:carbohydrate binding; C:integral component of membrane; C:anchored component of membrane | P:cellulose microfibril organization; C:anchored component of membrane | view details | |||
| JG647101.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 787 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650897.1 | Tinospora cordifolia | elongation factor 1-alpha-like | 827 | P:GO:0006414; P:GO:0090377; P:GO:0090378; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0043231 | P:translational elongation; P:seed trichome initiation; P:seed trichome elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG648137.1 | Tinospora cordifolia | probable inactive receptor kinase At4g23740 | 829 | P:GO:0006468; F:GO:0004672; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JG646742.1 | Tinospora cordifolia | 7-deoxyloganetin glucosyltransferase-like | 835 | F:GO:0080043; F:GO:0080044 | F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Glycosyltransferases | no GO terms | view details | ||
| JG650077.1 | Tinospora cordifolia | UTP--glucose-1-phosphate uridylyltransferase | 730 | P:GO:0006011; F:GO:0003983; C:GO:0016021 | P:UDP-glucose metabolic process; F:UTP:glucose-1-phosphate uridylyltransferase activity; C:integral component of membrane | UTP-monosaccharide-1-phosphate uridylyltransferase; UTP--glucose-1-phosphate uridylyltransferase | P:UDP-glucose metabolic process; F:UTP:glucose-1-phosphate uridylyltransferase activity; F:uridylyltransferase activity | view details | ||
| JG649282.1 | Tinospora cordifolia | basic endochitinase | 786 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650443.1 | Tinospora cordifolia | BURP domain protein RD22-like | 753 | no GO terms | view details | |||||
| JG646202.1 | Tinospora cordifolia | ADP-ribosylation factor 2 | 433 | P:GO:0015031; P:GO:0016192; F:GO:0003924; F:GO:0005525; C:GO:0005795 | P:protein transport; P:vesicle-mediated transport; F:GTPase activity; F:GTP binding; C:Golgi stack | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| JG648168.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 799 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647419.1 | Tinospora cordifolia | pentatricopeptide repeat-containing protein At3g53360, mitochondrial-like | 753 | P:GO:0009451; F:GO:0003723 | P:RNA modification; F:RNA binding | P:RNA modification; F:RNA binding; F:protein binding | view details | |||
| JG647128.1 | Tinospora cordifolia | heat shock 70 kDa protein 15-like | 750 | P:GO:0006457; F:GO:0005524; F:GO:0016887; C:GO:0005634; C:GO:0005788; C:GO:0005829 | P:protein folding; F:ATP binding; F:ATP hydrolysis activity; C:nucleus; C:endoplasmic reticulum lumen; C:cytosol | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG650122.1 | Tinospora cordifolia | basic endochitinase-like | 830 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647060.1 | Tinospora cordifolia | UPF0496 protein 4-like | 853 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| LQ667395.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1581 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG647397.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1 | 678 | F:GO:0003824 | F:catalytic activity | no GO terms | view details | |||
| JG645800.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 739 | no GO terms | view details | |||||
| JG646935.1 | Tinospora cordifolia | serine carboxypeptidase II-2 | 643 | F:GO:0004185; C:GO:0016020 | F:serine-type carboxypeptidase activity; C:membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type carboxypeptidase activity | view details | ||
| JG647772.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 747 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG650729.1 | Tinospora cordifolia | peroxidase P7-like | 807 | P:GO:0006979; P:GO:0042742; P:GO:0042744; P:GO:0098869; F:GO:0004601; F:GO:0020037; F:GO:0046872; C:GO:0005576; C:GO:0005737; C:GO:0009519; C:GO:0009531 | P:response to oxidative stress; P:defense response to bacterium; P:hydrogen peroxide catabolic process; P:cellular oxidant detoxification; F:peroxidase activity; F:heme binding; F:metal ion binding; C:extracellular region; C:cytoplasm; C:middle lamella; C:secondary cell wall | Acting on a peroxide as acceptor | P:response to oxidative stress; P:hydrogen peroxide catabolic process; F:peroxidase activity; F:heme binding | view details | ||
| JG647543.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 723 | F:GO:0050328; F:GO:0071949; C:GO:0005576 | F:tetrahydroberberine oxidase activity; F:FAD binding; C:extracellular region | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649981.1 | Tinospora cordifolia | cucumber peeling cupredoxin-like | 438 | C:GO:0046658 | C:anchored component of plasma membrane | F:electron transfer activity | view details | |||
| JG650252.1 | Tinospora cordifolia | hypersensitive-induced reaction 1 protein | 775 | C:GO:0005737; C:GO:0043231 | C:cytoplasm; C:intracellular membrane-bounded organelle | no GO terms | view details | |||
| JG647378.1 | Tinospora cordifolia | basic endochitinase-like | 772 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG649444.1 | Tinospora cordifolia | probable kinetochore protein SPC25 | 772 | P:GO:0007049; P:GO:0007059; P:GO:0051301; C:GO:0005634; C:GO:0031262 | P:cell cycle; P:chromosome segregation; P:cell division; C:nucleus; C:Ndc80 complex | P:chromosome segregation; C:Ndc80 complex | view details | |||
| JG650611.1 | Tinospora cordifolia | monodehydroascorbate reductase | 787 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG650903.1 | Tinospora cordifolia | V-type proton ATPase subunit B2-like | 130 | P:GO:0006811; F:GO:0005524 | P:ion transport; F:ATP binding | no GO terms | view details | |||
| JG646509.1 | Tinospora cordifolia | protein AAR2 homolog isoform X2 | 755 | P:GO:0000244; C:GO:0016021 | P:spliceosomal tri-snRNP complex assembly; C:integral component of membrane | no GO terms | view details | |||
| JG649996.1 | Tinospora cordifolia | serine carboxypeptidase II-2 | 820 | P:GO:0006508; F:GO:0004185; C:GO:0016020 | P:proteolysis; F:serine-type carboxypeptidase activity; C:membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:serine-type carboxypeptidase activity | view details | ||
| MS688299.1 | Tinospora cordifolia | RS-norcoclaurine 6-O-methyltransferase-like protein | 1065 | P:GO:0008152; F:GO:0008757 | P:metabolic process; F:S-adenosylmethionine-dependent methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| LQ667241.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 966 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| LQ667240.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 990 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG647516.1 | Tinospora cordifolia | transketolase, chloroplastic | 757 | P:GO:0006098; F:GO:0004802; F:GO:0046872; C:GO:0005829 | P:pentose-phosphate shunt; F:transketolase activity; F:metal ion binding; C:cytosol | Transketolase | no GO terms | view details | ||
| JG646950.1 | Tinospora cordifolia | 60S ribosomal protein L35a-1 | 214 | P:GO:0002181; P:GO:0042273; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; P:ribosomal large subunit biogenesis; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | no IPS match | view details | |||
| JG650726.1 | Tinospora cordifolia | glutathione S-transferase U17-like | 744 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG650997.1 | Tinospora cordifolia | Corytuberine synthase | 766 | P:GO:0009058; P:GO:0071704; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016020; C:GO:0043231 | P:biosynthetic process; P:organic substance metabolic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:membrane; C:intracellular membrane-bounded organelle | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648552.1 | Tinospora cordifolia | vesicle-fusing ATPase-like | 839 | P:GO:0006891; P:GO:0035494; P:GO:0043001; F:GO:0005524; F:GO:0016887; F:GO:0046872; C:GO:0005795 | P:intra-Golgi vesicle-mediated transport; P:SNARE complex disassembly; P:Golgi to plasma membrane protein transport; F:ATP binding; F:ATP hydrolysis activity; F:metal ion binding; C:Golgi stack | Acting on acid anhydrides | P:SNARE complex disassembly; F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG649401.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 787 | no GO terms | view details | |||||
| JG649782.1 | Tinospora cordifolia | basic endochitinase | 796 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647376.1 | Tinospora cordifolia | tRNA pseudouridine synthase A isoform X1 | 731 | P:GO:0031119; F:GO:0003723; F:GO:0009982 | P:tRNA pseudouridine synthesis; F:RNA binding; F:pseudouridine synthase activity | Intramolecular transferases | P:pseudouridine synthesis; P:RNA modification; F:RNA binding; F:pseudouridine synthase activity | view details | ||
| JG648799.1 | Tinospora cordifolia | kunitz trypsin inhibitor 5-like | 496 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646803.1 | Tinospora cordifolia | haloacid dehalogenase-like hydrolase domain-containing protein 3 | 446 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| JG649892.1 | Tinospora cordifolia | plant UBX domain-containing protein 11 | 253 | no GO terms | view details | |||||
| JG647662.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 840 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| MA258230.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1581 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648530.1 | Tinospora cordifolia | eukaryotic translation initiation factor 3 subunit A-like | 797 | P:GO:0001732; P:GO:0002188; F:GO:0003697; F:GO:0003729; F:GO:0003743; C:GO:0000502; C:GO:0016021; C:GO:0016282; C:GO:0033290; C:GO:0043614; C:GO:0071540; C:GO:0071541 | P:formation of cytoplasmic translation initiation complex; P:translation reinitiation; F:single-stranded DNA binding; F:mRNA binding; F:translation initiation factor activity; C:proteasome complex; C:integral component of membrane; C:eukaryotic 43S preinitiation complex; C:eukaryotic 48S preinitiation complex; C:multi-eIF complex; C:eukaryotic translation initiation factor 3 complex, eIF3e; C:eukaryotic translation initiation factor 3 complex, eIF3m | C:eukaryotic translation initiation factor 3 complex | view details | |||
| JG647902.1 | Tinospora cordifolia | methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | 760 | P:GO:0006210; P:GO:0006574; F:GO:0004491; F:GO:0018478; C:GO:0005739 | P:thymine catabolic process; P:valine catabolic process; F:methylmalonate-semialdehyde dehydrogenase (acylating) activity; F:malonate-semialdehyde dehydrogenase (acetylating) activity; C:mitochondrion | Methylmalonate-semialdehyde dehydrogenase (CoA acylating); Malonate-semialdehyde dehydrogenase (acetylating) | F:methylmalonate-semialdehyde dehydrogenase (acylating) activity; F:oxidoreductase activity | view details | ||
| JG649269.1 | Tinospora cordifolia | elongation factor 1-delta 1-like | 654 | P:GO:0006414; P:GO:0050790; F:GO:0003746; F:GO:0005085; C:GO:0005829; C:GO:0005853 | P:translational elongation; P:regulation of catalytic activity; F:translation elongation factor activity; F:guanyl-nucleotide exchange factor activity; C:cytosol; C:eukaryotic translation elongation factor 1 complex | P:translational elongation; F:translation elongation factor activity | view details | |||
| JG647444.1 | Tinospora cordifolia | glucose-6-phosphate 1-dehydrogenase, cytoplasmic isoform | 756 | P:GO:0006006; P:GO:0009051; F:GO:0004345; F:GO:0050661; C:GO:0005829 | P:glucose metabolic process; P:pentose-phosphate shunt, oxidative branch; F:glucose-6-phosphate dehydrogenase activity; F:NADP binding; C:cytosol | Glucose-6-phosphate dehydrogenase (NADP(+)) | P:glucose metabolic process; F:oxidoreductase activity, acting on CH-OH group of donors; F:NADP binding | view details | ||
| JG648852.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 857 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646291.1 | Tinospora cordifolia | berberine bridge enzyme-like 8 | 726 | F:GO:0016491; F:GO:0071949; C:GO:0016021 | F:oxidoreductase activity; F:FAD binding; C:integral component of membrane | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650987.1 | Tinospora cordifolia | glucosamine inositolphosphorylceramide transferase 1-like | 778 | P:GO:0006486; F:GO:0016757; C:GO:0016021 | P:protein glycosylation; F:glycosyltransferase activity; C:integral component of membrane | Glycosyltransferases | P:protein glycosylation; F:glycosyltransferase activity | view details | ||
| JG649592.1 | Tinospora cordifolia | probable pectinesterase/pectinesterase inhibitor 7 | 835 | F:GO:0030599; F:GO:0046910 | F:pectinesterase activity; F:pectinesterase inhibitor activity | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG649325.1 | Tinospora cordifolia | Fad-binding berberine family protein | 753 | F:GO:0016491; F:GO:0071949; C:GO:0005576 | F:oxidoreductase activity; F:FAD binding; C:extracellular region | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648906.1 | Tinospora cordifolia | auxin response factor 2A-like | 839 | P:GO:0009725; P:GO:0050794 | P:response to hormone; P:regulation of cellular process | P:regulation of transcription, DNA-templated; P:response to hormone; F:DNA binding | view details | |||
| JG645837.1 | Tinospora cordifolia | pleiotropic drug resistance protein 1-like | 438 | P:GO:0002229; P:GO:0009408; P:GO:0009409; P:GO:0009738; P:GO:0010193; P:GO:0010496; P:GO:0015692; P:GO:0042631; P:GO:0048581; P:GO:0080168; P:GO:0090332; P:GO:0098657; P:GO:0098739; F:GO:0005524; F:GO:0015562; F:GO:0140359; C:GO:0005886; C:GO:0016021 | P:defense response to oomycetes; P:response to heat; P:response to cold; P:abscisic acid-activated signaling pathway; P:response to ozone; P:intercellular transport; P:lead ion transport; P:cellular response to water deprivation; P:negative regulation of post-embryonic development; P:abscisic acid transport; P:stomatal closure; P:import into cell; P:import across plasma membrane; F:ATP binding; F:efflux transmembrane transporter activity; F:ABC-type transporter activity; C:plasma membrane; C:integral component of membrane | Translocases | C:membrane | view details | ||
| JG650675.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 849 | F:GO:0005524; F:GO:0016887 | F:ATP binding; F:ATP hydrolysis activity | Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG647671.1 | Tinospora cordifolia | cytochrome p450 82c4 | 799 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648516.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 759 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG647449.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 777 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650345.1 | Tinospora cordifolia | thaumatin-like protein | 574 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650789.1 | Tinospora cordifolia | actin-101 | 864 | F:GO:0005524; C:GO:0005737; C:GO:0005856 | F:ATP binding; C:cytoplasm; C:cytoskeleton | no GO terms | view details | |||
| JG646113.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 808 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647020.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 844 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649921.1 | Tinospora cordifolia | 40S ribosomal protein S15a-1 | 191 | P:GO:0006412; F:GO:0003735; C:GO:0005840 | P:translation; F:structural constituent of ribosome; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649019.1 | Tinospora cordifolia | ---NA--- | 118 | no IPS match | view details | |||||
| JG648566.1 | Tinospora cordifolia | putative galacturonosyltransferase 14 | 752 | P:GO:0045489; P:GO:0071555; F:GO:0047262; C:GO:0000139; C:GO:0016021 | P:pectin biosynthetic process; P:cell wall organization; F:polygalacturonate 4-alpha-galacturonosyltransferase activity; C:Golgi membrane; C:integral component of membrane | Polygalacturonate 4-alpha-galacturonosyltransferase | F:glycosyltransferase activity; F:polygalacturonate 4-alpha-galacturonosyltransferase activity | view details | ||
| JG649165.1 | Tinospora cordifolia | BURP domain protein RD22-like | 835 | no GO terms | view details | |||||
| JG649240.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 706 | no GO terms | view details | |||||
| JG647161.1 | Tinospora cordifolia | formin-like protein 5 | 817 | P:GO:0045010; F:GO:0051015; C:GO:0016021 | P:actin nucleation; F:actin filament binding; C:integral component of membrane | P:actin cytoskeleton organization; P:actin nucleation; F:actin binding; F:actin filament binding | view details | |||
| JG647071.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 819 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG648969.1 | Tinospora cordifolia | 5-oxoprolinase-like | 431 | F:GO:0016787 | F:hydrolase activity | Hydrolases | F:hydrolase activity | view details | ||
| JG649579.1 | Tinospora cordifolia | glucan endo-1,3-beta-D-glucosidase-like | 814 | P:GO:0005975; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650876.1 | Tinospora cordifolia | alpha-dioxygenase 1-like | 829 | P:GO:0006979; P:GO:0098869; F:GO:0004601; F:GO:0016702; F:GO:0020037 | P:response to oxidative stress; P:cellular oxidant detoxification; F:peroxidase activity; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen; F:heme binding | Acting on single donors with incorporation of molecular oxygen (oxygenases). The oxygen incorporated need not be derived from O(2); Acting on a peroxide as acceptor | P:response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG647910.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 776 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG650643.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 832 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG647345.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 779 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650689.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 883 | P:GO:0005975; P:GO:0006952; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645979.1 | Tinospora cordifolia | (-)-alpha-pinene synthase-like | 814 | P:GO:0016114; F:GO:0010333; F:GO:0046872 | P:terpenoid biosynthetic process; F:terpene synthase activity; F:metal ion binding | Carbon-oxygen lyases | F:magnesium ion binding; F:terpene synthase activity; F:lyase activity | view details | ||
| JG649020.1 | Tinospora cordifolia | 40S ribosomal protein S19-1-like | 274 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; F:GO:0016740; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; F:transferase activity; C:cytosolic small ribosomal subunit | Transferases | P:translation; F:structural constituent of ribosome; C:ribosome | view details | ||
| JG646308.1 | Tinospora cordifolia | ethylene receptor isoform X1 | 780 | P:GO:0010105; P:GO:0018106; P:GO:0048856; F:GO:0000155; F:GO:0005524; F:GO:0038199; F:GO:0046872; F:GO:0051740; C:GO:0005789; C:GO:0016021 | P:negative regulation of ethylene-activated signaling pathway; P:peptidyl-histidine phosphorylation; P:anatomical structure development; F:phosphorelay sensor kinase activity; F:ATP binding; F:ethylene receptor activity; F:metal ion binding; F:ethylene binding; C:endoplasmic reticulum membrane; C:integral component of membrane | Transferring phosphorus-containing groups; Histidine kinase | F:protein binding | view details | ||
| JG646838.1 | Tinospora cordifolia | probable glutathione S-transferase | 610 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG646914.1 | Tinospora cordifolia | 50S ribosomal protein L27, chloroplastic | 504 | P:GO:0006412; F:GO:0003735; C:GO:0005737; C:GO:0005840 | P:translation; F:structural constituent of ribosome; C:cytoplasm; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648122.1 | Tinospora cordifolia | monodehydroascorbate reductase | 774 | P:GO:0098869; F:GO:0016656; F:GO:0050660; C:GO:0005737 | P:cellular oxidant detoxification; F:monodehydroascorbate reductase (NADH) activity; F:flavin adenine dinucleotide binding; C:cytoplasm | Monodehydroascorbate reductase (NADH) | F:oxidoreductase activity | view details | ||
| JG645538.1 | Tinospora cordifolia | Oxysterol-binding protein-related protein 1d | 730 | P:GO:0015918; F:GO:0015248; F:GO:0032934; C:GO:0005829; C:GO:0016020; C:GO:0043231 | P:sterol transport; F:sterol transporter activity; F:sterol binding; C:cytosol; C:membrane; C:intracellular membrane-bounded organelle | Translocases | F:lipid binding | view details | ||
| JG649954.1 | Tinospora cordifolia | thaumatin-like protein | 719 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG648037.1 | Tinospora cordifolia | NADPH-dependent aldo-keto reductase, chloroplastic | 750 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG646760.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 796 | P:GO:0005975; P:GO:0006952; F:GO:0004553; F:GO:0016765; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647620.1 | Tinospora cordifolia | 4-alpha-glucanotransferase DPE2 | 727 | P:GO:0005977; F:GO:0004134; F:GO:0016787; F:GO:0102500; F:GO:2001070 | P:glycogen metabolic process; F:4-alpha-glucanotransferase activity; F:hydrolase activity; F:beta-maltose 4-alpha-glucanotransferase activity; F:starch binding | 4-alpha-glucanotransferase; Hydrolases | P:carbohydrate metabolic process; F:4-alpha-glucanotransferase activity | view details | ||
| JG650910.1 | Tinospora cordifolia | Conserved oligomeric Golgi complex subunit 6 | 718 | P:GO:0006891; P:GO:0015031; C:GO:0000139; C:GO:0017119 | P:intra-Golgi vesicle-mediated transport; P:protein transport; C:Golgi membrane; C:Golgi transport complex | P:intra-Golgi vesicle-mediated transport; C:Golgi transport complex | view details | |||
| JG647251.1 | Tinospora cordifolia | basic endochitinase-like | 816 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| KT384103.1 | Tinospora cordifolia | protein WHAT S THIS FACTOR 1 homolog, chloroplastic | 228 | F:GO:0003723; F:GO:0016787 | F:RNA binding; F:hydrolase activity | Hydrolases | F:RNA binding | view details | ||
| JG650175.1 | Tinospora cordifolia | Acyltransferase-like protein | 588 | P:GO:0009820; F:GO:0004144 | P:alkaloid metabolic process; F:diacylglycerol O-acyltransferase activity | Diacylglycerol O-acyltransferase | no GO terms | view details | ||
| JG646492.1 | Tinospora cordifolia | calmodulin-binding protein 60 D-like isoform X3 | 407 | P:GO:0006355; P:GO:0080142; F:GO:0003700; F:GO:0005516; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:regulation of salicylic acid biosynthetic process; F:DNA-binding transcription factor activity; F:calmodulin binding; F:sequence-specific DNA binding; C:nucleus | F:calmodulin binding | view details | |||
| JG649854.1 | Tinospora cordifolia | BURP domain protein RD22-like | 747 | no GO terms | view details | |||||
| JG648881.1 | Tinospora cordifolia | S-adenosylmethionine synthase | 842 | P:GO:0006556; P:GO:0006730; F:GO:0004478; F:GO:0005524; F:GO:0046872; C:GO:0005829 | P:S-adenosylmethionine biosynthetic process; P:one-carbon metabolic process; F:methionine adenosyltransferase activity; F:ATP binding; F:metal ion binding; C:cytosol | Methionine adenosyltransferase | P:S-adenosylmethionine biosynthetic process; F:methionine adenosyltransferase activity; F:ATP binding | view details | ||
| JG645524.1 | Tinospora cordifolia | eukaryotic translation initiation factor-like | 738 | P:GO:0006413; P:GO:0006417; F:GO:0003729; F:GO:0003743; C:GO:0016281 | P:translational initiation; P:regulation of translation; F:mRNA binding; F:translation initiation factor activity; C:eukaryotic translation initiation factor 4F complex | F:RNA binding; F:translation initiation factor activity; F:protein binding | view details | |||
| JG649196.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 561 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG647966.1 | Tinospora cordifolia | casein kinase II subunit alpha-like | 777 | P:GO:0018105; P:GO:0018107; P:GO:0051726; F:GO:0004674; F:GO:0005524 | P:peptidyl-serine phosphorylation; P:peptidyl-threonine phosphorylation; P:regulation of cell cycle; F:protein serine/threonine kinase activity; F:ATP binding | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:protein serine/threonine kinase activity; F:ATP binding | view details | ||
| JG649496.1 | Tinospora cordifolia | cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like | 745 | P:GO:0006122; P:GO:1902600; F:GO:0008121; F:GO:0046872; F:GO:0051537; C:GO:0005750 | P:mitochondrial electron transport, ubiquinol to cytochrome c; P:proton transmembrane transport; F:ubiquinol-cytochrome-c reductase activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; C:mitochondrial respiratory chain complex III | Acting on diphenols and related substances as donors; Quinol--cytochrome-c reductase | F:ubiquinol-cytochrome-c reductase activity; F:2 iron, 2 sulfur cluster binding; C:membrane | view details | ||
| JG646808.1 | Tinospora cordifolia | cytochrome P450 94B3-like | 773 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650594.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 861 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG645882.1 | Tinospora cordifolia | tetratricopeptide repeat protein 38 | 835 | F:protein binding | view details | |||||
| JG647537.1 | Tinospora cordifolia | ATP-dependent RNA helicase DEAH11, chloroplastic-like | 687 | P:GO:0044238; P:GO:0044260; F:GO:0003723; F:GO:0003724; F:GO:0005524; F:GO:0016740; F:GO:0016787; F:GO:0046872 | P:primary metabolic process; P:cellular macromolecule metabolic process; F:RNA binding; F:RNA helicase activity; F:ATP binding; F:transferase activity; F:hydrolase activity; F:metal ion binding | RNA helicase; Transferases | P:protein ubiquitination; F:ubiquitin-protein transferase activity | view details | ||
| JG648966.1 | Tinospora cordifolia | ---NA--- | 116 | no GO terms | view details | |||||
| JG649830.1 | Tinospora cordifolia | probable quinone oxidoreductase | 772 | P:GO:0006979; F:GO:0008270; F:GO:0016491 | P:response to oxidative stress; F:zinc ion binding; F:oxidoreductase activity | Oxidoreductases | no GO terms | view details | ||
| JG648743.1 | Tinospora cordifolia | (6-4)DNA photolyase isoform X2 | 763 | P:GO:0006290; P:GO:0009411; P:GO:0032922; P:GO:0043153; F:GO:0003677; F:GO:0003914; F:GO:0071949; C:GO:0005634; C:GO:0005737; C:GO:0016021 | P:pyrimidine dimer repair; P:response to UV; P:circadian regulation of gene expression; P:entrainment of circadian clock by photoperiod; F:DNA binding; F:DNA (6-4) photolyase activity; F:FAD binding; C:nucleus; C:cytoplasm; C:integral component of membrane | (6-4)DNA photolyase | no GO terms | view details | ||
| JG646393.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 855 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649441.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 839 | F:GO:0050660 | F:flavin adenine dinucleotide binding | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | |||
| JG647809.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 761 | P:GO:0005975; P:GO:0006952; F:GO:0008061; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; P:defense response; F:chitin binding; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649076.1 | Tinospora cordifolia | mogroside IE synthase-like | 719 | F:GO:0016757 | F:glycosyltransferase activity | Glycosyltransferases | no GO terms | view details | ||
| JG649528.1 | Tinospora cordifolia | cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like | 830 | P:GO:0006122; P:GO:1902600; F:GO:0008121; F:GO:0046872; F:GO:0051537; C:GO:0005750 | P:mitochondrial electron transport, ubiquinol to cytochrome c; P:proton transmembrane transport; F:ubiquinol-cytochrome-c reductase activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; C:mitochondrial respiratory chain complex III | Acting on diphenols and related substances as donors; Quinol--cytochrome-c reductase | F:ubiquinol-cytochrome-c reductase activity; F:2 iron, 2 sulfur cluster binding; C:membrane | view details | ||
| JG650236.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 769 | F:GO:0050328; F:GO:0071949; C:GO:0016020 | F:tetrahydroberberine oxidase activity; F:FAD binding; C:membrane | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648233.1 | Tinospora cordifolia | pectinesterase/pectinesterase inhibitor PPE8B | 647 | P:GO:0071555; F:GO:0030599; F:GO:0046910 | P:cell wall organization; F:pectinesterase activity; F:pectinesterase inhibitor activity | Pectinesterase | P:cell wall modification; F:enzyme inhibitor activity; F:pectinesterase activity | view details | ||
| JG646725.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP) | 742 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG645859.1 | Tinospora cordifolia | laccase-7-like | 891 | P:GO:0046274; F:GO:0005507; F:GO:0052716; C:GO:0048046 | P:lignin catabolic process; F:copper ion binding; F:hydroquinone:oxygen oxidoreductase activity; C:apoplast | Laccase | F:copper ion binding | view details | ||
| JG650223.1 | Tinospora cordifolia | protein unc-13 homolog | 807 | no GO terms | view details | |||||
| JG646538.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1-like | 729 | F:GO:0003824 | F:catalytic activity | no GO terms | view details | |||
| JG649613.1 | Tinospora cordifolia | ATP synthase subunit gamma, mitochondrial | 721 | P:GO:0006811; P:GO:0015986; F:GO:0046933; C:GO:0000275 | P:ion transport; P:proton motive force-driven ATP synthesis; F:proton-transporting ATP synthase activity, rotational mechanism; C:mitochondrial proton-transporting ATP synthase complex, catalytic sector F(1) | H(+)-transporting two-sector ATPase; Ligases | P:proton motive force-driven ATP synthesis; F:proton-transporting ATP synthase activity, rotational mechanism; C:proton-transporting ATP synthase complex, catalytic core F(1) | view details | ||
| JG650899.1 | Tinospora cordifolia | dnaJ protein homolog | 771 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG647941.1 | Tinospora cordifolia | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase-like | 740 | P:GO:0009086; P:GO:0032259; F:GO:0003871; F:GO:0008270; F:GO:0008705; C:GO:0005829; C:GO:0009507 | P:methionine biosynthetic process; P:methylation; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding; F:methionine synthase activity; C:cytosol; C:chloroplast | Methionine synthase; 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase | P:cellular amino acid biosynthetic process; F:5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity; F:zinc ion binding | view details | ||
| JG646949.1 | Tinospora cordifolia | ---NA--- | 79 | no IPS match | view details | |||||
| JG646752.1 | Tinospora cordifolia | berberine bridge enzyme-like 18 | 826 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG650458.1 | Tinospora cordifolia | basic endochitinase-like | 831 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG647198.1 | Tinospora cordifolia | CBL-interacting serine/threonine-protein kinase 14 | 837 | P:GO:0006468; P:GO:0007165; F:GO:0004674; F:GO:0004712; F:GO:0005524; F:GO:0106310; C:GO:0016021 | P:protein phosphorylation; P:signal transduction; F:protein serine/threonine kinase activity; F:protein serine/threonine/tyrosine kinase activity; F:ATP binding; F:protein serine kinase activity; C:integral component of membrane | Dual-specificity kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG650332.1 | Tinospora cordifolia | methylecgonone reductase | 797 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG650363.1 | Tinospora cordifolia | probable pectinesterase/pectinesterase inhibitor 7 | 823 | F:GO:0030599; F:GO:0046910; C:GO:0016020 | F:pectinesterase activity; F:pectinesterase inhibitor activity; C:membrane | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG647931.1 | Tinospora cordifolia | succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial-like | 587 | P:GO:0006099; P:GO:0006124; P:GO:0022904; F:GO:0008177; F:GO:0009055; F:GO:0046872; F:GO:0051537; F:GO:0051538; F:GO:0051539; C:GO:0005743; C:GO:0009507; C:GO:0045273 | P:tricarboxylic acid cycle; P:ferredoxin metabolic process; P:respiratory electron transport chain; F:succinate dehydrogenase (ubiquinone) activity; F:electron transfer activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; F:3 iron, 4 sulfur cluster binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrial inner membrane; C:chloroplast; C:respiratory chain complex II | Succinate dehydrogenase (quinone) | P:tricarboxylic acid cycle; F:electron transfer activity; F:oxidoreductase activity; F:iron-sulfur cluster binding | view details | ||
| JG647896.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 776 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG649531.1 | Tinospora cordifolia | 7-deoxyloganetin glucosyltransferase-like | 842 | F:GO:0080043; F:GO:0080044 | F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Glycosyltransferases | no GO terms | view details | ||
| JG648937.1 | Tinospora cordifolia | 2-alkenal reductase (NADP(+)-dependent)-like | 813 | F:GO:0016628 | F:oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | Acting on the CH-CH group of donors | F:oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | view details | ||
| JG650468.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase-like | 812 | F:GO:0016491; F:GO:0046872 | F:oxidoreductase activity; F:metal ion binding | Oxidoreductases | no GO terms | view details | ||
| MA258131.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 960 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG646814.1 | Tinospora cordifolia | polygalacturonase-inhibiting protein | 774 | C:GO:0110165 | C:cellular anatomical entity | F:protein binding | view details | |||
| JG649869.1 | Tinospora cordifolia | basic endochitinase | 798 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646554.1 | Tinospora cordifolia | EIN3-binding F-box protein 1-like | 773 | P:GO:0031146; C:GO:0019005 | P:SCF-dependent proteasomal ubiquitin-dependent protein catabolic process; C:SCF ubiquitin ligase complex | F:protein binding | view details | |||
| JG648577.1 | Tinospora cordifolia | elongation factor 1-alpha-like | 739 | P:GO:0006414; P:GO:0090377; P:GO:0090378; F:GO:0003746; F:GO:0003924; F:GO:0005525; C:GO:0005737; C:GO:0043231 | P:translational elongation; P:seed trichome initiation; P:seed trichome elongation; F:translation elongation factor activity; F:GTPase activity; F:GTP binding; C:cytoplasm; C:intracellular membrane-bounded organelle | Acting on acid anhydrides | F:GTPase activity; F:GTP binding | view details | ||
| MA258232.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 1596 | P:GO:0009058; P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:biosynthetic process; P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646551.1 | Tinospora cordifolia | Berberine bridge enzyme-like 8 | 701 | F:GO:0050328; F:GO:0071949; C:GO:0005576 | F:tetrahydroberberine oxidase activity; F:FAD binding; C:extracellular region | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| LY340440.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 1581 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649303.1 | Tinospora cordifolia | ---NA--- | 104 | no IPS match | view details | |||||
| JG645724.1 | Tinospora cordifolia | transformation/transcription domain-associated protein-like | 838 | P:GO:0006281; P:GO:0006355; P:GO:0006468; F:GO:0003712; F:GO:0004674; C:GO:0000124; C:GO:0035267 | P:DNA repair; P:regulation of transcription, DNA-templated; P:protein phosphorylation; F:transcription coregulator activity; F:protein serine/threonine kinase activity; C:SAGA complex; C:NuA4 histone acetyltransferase complex | Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JG646937.1 | Tinospora cordifolia | ---NA--- | 66 | no GO terms | view details | |||||
| JG647318.1 | Tinospora cordifolia | probable methyltransferase PMT21 | 794 | P:GO:0032259; F:GO:0008168; C:GO:0005737; C:GO:0016021; C:GO:0043231 | P:methylation; F:methyltransferase activity; C:cytoplasm; C:integral component of membrane; C:intracellular membrane-bounded organelle | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG648011.1 | Tinospora cordifolia | nitrate reductase [NADH]-like | 846 | P:GO:0006809; P:GO:0042128; F:GO:0009703; F:GO:0020037; F:GO:0030151; F:GO:0043546; F:GO:0050464; F:GO:0071949 | P:nitric oxide biosynthetic process; P:nitrate assimilation; F:nitrate reductase (NADH) activity; F:heme binding; F:molybdenum ion binding; F:molybdopterin cofactor binding; F:nitrate reductase (NADPH) activity; F:FAD binding | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Nitrate reductase (NADPH); Nitrate reductase | F:oxidoreductase activity; F:molybdenum ion binding | view details | ||
| JG647905.1 | Tinospora cordifolia | S-adenosyl-L-methionine:norcoclaurine 6-O-methyltransferase | 852 | P:GO:0009821; P:GO:0019438; P:GO:0032259; F:GO:0008171; F:GO:0030786; F:GO:0042802; F:GO:0046983 | P:alkaloid biosynthetic process; P:aromatic compound biosynthetic process; P:methylation; F:O-methyltransferase activity; F:(RS)-norcoclaurine 6-O-methyltransferase activity; F:identical protein binding; F:protein dimerization activity | (RS)-norcoclaurine 6-O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG650258.1 | Tinospora cordifolia | EEF1A lysine methyltransferase 2-like | 745 | P:GO:0018026; P:GO:0018027; F:GO:0016279; C:GO:0005737 | P:peptidyl-lysine monomethylation; P:peptidyl-lysine dimethylation; F:protein-lysine N-methyltransferase activity; C:cytoplasm | Transferring one-carbon groups | no GO terms | view details | ||
| JG647138.1 | Tinospora cordifolia | auxin-responsive protein IAA9-like isoform X1 | 771 | P:GO:0006355; P:GO:0009734; F:GO:0016740; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:auxin-activated signaling pathway; F:transferase activity; C:nucleus | P:regulation of transcription, DNA-templated; F:protein binding; C:nucleus | view details | |||
| JG646218.1 | Tinospora cordifolia | pectinesterase 2 | 627 | P:GO:0042545; P:GO:0043086; P:GO:0045490; F:GO:0030599; F:GO:0045330; F:GO:0046910; C:GO:0005576 | P:cell wall modification; P:negative regulation of catalytic activity; P:pectin catabolic process; F:pectinesterase activity; F:aspartyl esterase activity; F:pectinesterase inhibitor activity; C:extracellular region | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG645957.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 832 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG650357.1 | Tinospora cordifolia | protein SOMBRERO isoform X1 | 833 | P:GO:0003002; P:GO:0006355; P:GO:0009834; P:GO:0010455; P:GO:0048829; F:GO:0003677; C:GO:0005634 | P:regionalization; P:regulation of transcription, DNA-templated; P:plant-type secondary cell wall biogenesis; P:positive regulation of cell fate commitment; P:root cap development; F:DNA binding; C:nucleus | P:regulation of transcription, DNA-templated; F:DNA binding | view details | |||
| JG649188.1 | Tinospora cordifolia | phosphoserine aminotransferase 2, chloroplastic-like | 758 | P:GO:0006564; F:GO:0004648; F:GO:0030170; C:GO:0005737 | P:L-serine biosynthetic process; F:O-phospho-L-serine:2-oxoglutarate aminotransferase activity; F:pyridoxal phosphate binding; C:cytoplasm | Phosphoserine transaminase | P:L-serine biosynthetic process; F:catalytic activity; F:O-phospho-L-serine:2-oxoglutarate aminotransferase activity | view details | ||
| JG648111.1 | Tinospora cordifolia | transmembrane protein 256 homolog | 327 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG649824.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 807 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG649032.1 | Tinospora cordifolia | Par1 protein | 132 | no IPS match | view details | |||||
| JG648603.1 | Tinospora cordifolia | SOUL heme-binding protein | 812 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| JG645919.1 | Tinospora cordifolia | pathogen-related protein-like | 738 | no GO terms | view details | |||||
| JG645691.1 | Tinospora cordifolia | zinc finger CCCH domain-containing protein 53-like isoform X1 | 829 | F:GO:0003677; F:GO:0003723; F:GO:0046872 | F:DNA binding; F:RNA binding; F:metal ion binding | F:nucleic acid binding; F:RNA binding | view details | |||
| JG650158.1 | Tinospora cordifolia | armadillo repeat-containing protein 8-like | 793 | P:GO:0043161; C:GO:0005634; C:GO:0005737; C:GO:0034657 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; C:nucleus; C:cytoplasm; C:GID complex | no GO terms | view details | |||
| JG649581.1 | Tinospora cordifolia | basic endochitinase-like | 603 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | no IPS match | view details | ||
| JG649557.1 | Tinospora cordifolia | plant UBX domain-containing protein 8-like isoform X2 | 590 | P:GO:0030433; F:GO:0043130; C:GO:0005783 | P:ubiquitin-dependent ERAD pathway; F:ubiquitin binding; C:endoplasmic reticulum | no GO terms | view details | |||
| JG645993.1 | Tinospora cordifolia | cystathionine gamma-synthase 1, chloroplastic-like | 810 | P:GO:0009086; P:GO:0019346; F:GO:0003962; F:GO:0030170 | P:methionine biosynthetic process; P:transsulfuration; F:cystathionine gamma-synthase activity; F:pyridoxal phosphate binding | Cystathionine gamma-synthase | P:methionine biosynthetic process; P:transsulfuration; F:catalytic activity; F:cystathionine gamma-synthase activity; F:pyridoxal phosphate binding | view details | ||
| JG647028.1 | Tinospora cordifolia | heat shock cognate 70 kDa protein 2 | 769 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0032440; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005737 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:2-alkenal reductase [NAD(P)+] activity; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:cytoplasm | 2-alkenal reductase (NAD(P)(+)); Acting on acid anhydrides | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG646030.1 | Tinospora cordifolia | NEDD8 ultimate buster 1 | 707 | P:GO:0006414; P:GO:2000058; F:GO:0003746; F:GO:0031593; C:GO:0005634 | P:translational elongation; P:regulation of ubiquitin-dependent protein catabolic process; F:translation elongation factor activity; F:polyubiquitin modification-dependent protein binding; C:nucleus | F:protein binding | view details | |||
| JG646072.1 | Tinospora cordifolia | Sulfate/thiosulfate import ATP-binding protein cysA | 811 | F:GO:0005524 | F:ATP binding | no GO terms | view details | |||
| JG646582.1 | Tinospora cordifolia | translation machinery-associated protein 22 | 253 | P:GO:0001731; P:GO:0002188; F:GO:0003729; F:GO:0003743; C:GO:0005737; C:GO:0005840 | P:formation of translation preinitiation complex; P:translation reinitiation; F:mRNA binding; F:translation initiation factor activity; C:cytoplasm; C:ribosome | no IPS match | view details | |||
| JG645789.1 | Tinospora cordifolia | golgin family A protein | 729 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG647930.1 | Tinospora cordifolia | WRKY transcription factor WRKY24 | 699 | P:GO:0006355; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG647065.1 | Tinospora cordifolia | pirin-like protein | 770 | no GO terms | view details | |||||
| JG649304.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 764 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG648932.1 | Tinospora cordifolia | E3 ubiquitin-protein ligase RNF185-like | 742 | P:GO:0006511; P:GO:0016310; P:GO:0016567; P:GO:0071712; F:GO:0016301; F:GO:0044390; F:GO:0046872; F:GO:0061630; C:GO:0005783; C:GO:0016021 | P:ubiquitin-dependent protein catabolic process; P:phosphorylation; P:protein ubiquitination; P:ER-associated misfolded protein catabolic process; F:kinase activity; F:ubiquitin-like protein conjugating enzyme binding; F:metal ion binding; F:ubiquitin protein ligase activity; C:endoplasmic reticulum; C:integral component of membrane | Transferring phosphorus-containing groups | P:ubiquitin-dependent protein catabolic process; F:metal ion binding; F:ubiquitin protein ligase activity; C:endoplasmic reticulum | view details | ||
| JG647597.1 | Tinospora cordifolia | receptor-like serine/threonine-protein kinase At1g78530 | 817 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0005886; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:plasma membrane; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG650707.1 | Tinospora cordifolia | COBW domain-containing protein 1 isoform X1 | 859 | C:GO:0005737 | C:cytoplasm | no GO terms | view details | |||
| JG650212.1 | Tinospora cordifolia | probable 1-acylglycerol-3-phosphate O-acyltransferase | 718 | P:GO:0006654; P:GO:0055088; F:GO:0004623; F:GO:0042171 | P:phosphatidic acid biosynthetic process; P:lipid homeostasis; F:phospholipase A2 activity; F:lysophosphatidic acid acyltransferase activity | Carboxylesterase; Phospholipase A(2); Acyltransferases | P:proteolysis; F:peptidase activity | view details | ||
| JG646883.1 | Tinospora cordifolia | EEIG1/EHBP1 N-terminal domain | 426 | no IPS match | view details | |||||
| JG646827.1 | Tinospora cordifolia | protein usf | 648 | F:GO:0008806 | F:carboxymethylenebutenolidase activity | Carboxymethylenebutenolidase; Carboxylesterase | F:hydrolase activity | view details | ||
| JG650253.1 | Tinospora cordifolia | protein HUA2-LIKE 3-like | 803 | P:GO:0006357; P:GO:0006397; F:GO:0003690; F:GO:0003712; C:GO:0005634 | P:regulation of transcription by RNA polymerase II; P:mRNA processing; F:double-stranded DNA binding; F:transcription coregulator activity; C:nucleus | no GO terms | view details | |||
| JG646284.1 | Tinospora cordifolia | glutathione S-transferase L3 | 622 | P:GO:0006749; P:GO:0010731; F:GO:0004364 | P:glutathione metabolic process; P:protein glutathionylation; F:glutathione transferase activity | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG649675.1 | Tinospora cordifolia | polyubiquitin 11 | 778 | P:GO:0006511; P:GO:0016567; F:GO:0003729; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:ubiquitin-dependent protein catabolic process; P:protein ubiquitination; F:mRNA binding; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm; C:ribosome | F:protein binding | view details | |||
| JG647623.1 | Tinospora cordifolia | CCR4-NOT transcription complex subunit 11 | 785 | P:GO:0006402; P:GO:0017148; P:GO:0031047; C:GO:0005634; C:GO:0005737; C:GO:0030014 | P:mRNA catabolic process; P:negative regulation of translation; P:gene silencing by RNA; C:nucleus; C:cytoplasm; C:CCR4-NOT complex | C:CCR4-NOT complex | view details | |||
| JG649748.1 | Tinospora cordifolia | Receptor like protein kinase S.2 | 657 | P:GO:0002229; P:GO:0016310; P:GO:0042742; F:GO:0000166; F:GO:0004675; C:GO:0005886 | P:defense response to oomycetes; P:phosphorylation; P:defense response to bacterium; F:nucleotide binding; F:transmembrane receptor protein serine/threonine kinase activity; C:plasma membrane | Receptor protein serine/threonine kinase | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG646929.1 | Tinospora cordifolia | Nucleoside phosphatase GDA1/CD39 | 635 | F:GO:0005524; F:GO:0016787; C:GO:0016021 | F:ATP binding; F:hydrolase activity; C:integral component of membrane | Hydrolases | F:hydrolase activity | view details | ||
| JG649307.1 | Tinospora cordifolia | probable transmembrane ascorbate ferrireductase 2 | 773 | P:GO:0019852; F:GO:0000293; C:GO:0016021 | P:L-ascorbic acid metabolic process; F:ferric-chelate reductase activity; C:integral component of membrane | Oxidizing metal ions | F:oxidoreductase activity | view details | ||
| JG645526.1 | Tinospora cordifolia | formate dehydrogenase, mitochondrial | 553 | P:GO:0042183; F:GO:0008863; F:GO:0016616; F:GO:0051287; C:GO:0005739; C:GO:0009326; C:GO:0009507 | P:formate catabolic process; F:formate dehydrogenase (NAD+) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding; C:mitochondrion; C:formate dehydrogenase complex; C:chloroplast | Acting on the CH-OH group of donors; Formate dehydrogenase; Acting on the aldehyde or oxo group of donors | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG648007.1 | Tinospora cordifolia | plant UBX domain-containing protein 8 | 707 | P:GO:0030433; F:GO:0043130; C:GO:0005783 | P:ubiquitin-dependent ERAD pathway; F:ubiquitin binding; C:endoplasmic reticulum | F:protein binding | view details | |||
| JG647508.1 | Tinospora cordifolia | pathogen-related protein-like | 733 | no GO terms | view details | |||||
| JG648072.1 | Tinospora cordifolia | probable inositol transporter 2 | 827 | P:GO:0006817; P:GO:0015798; P:GO:0050896; P:GO:0055085; F:GO:0005366; C:GO:0016021 | P:phosphate ion transport; P:myo-inositol transport; P:response to stimulus; P:transmembrane transport; F:myo-inositol:proton symporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG646278.1 | Tinospora cordifolia | Pyrimidine 5-nucleotidase protein | 699 | P:GO:0016311; F:GO:0033883 | P:dephosphorylation; F:pyridoxal phosphatase activity | Pyridoxal phosphatase | no GO terms | view details | ||
| JG650191.1 | Tinospora cordifolia | dnaJ homolog subfamily B member 1 | 812 | P:GO:0051085; F:GO:0043130; F:GO:0051082; F:GO:0051087; C:GO:0005829 | P:chaperone cofactor-dependent protein refolding; F:ubiquitin binding; F:unfolded protein binding; F:chaperone binding; C:cytosol | P:protein folding; F:unfolded protein binding | view details | |||
| JG647278.1 | Tinospora cordifolia | formate dehydrogenase, mitochondrial | 776 | P:GO:0042183; F:GO:0008863; F:GO:0016616; F:GO:0051287; C:GO:0005739; C:GO:0009326; C:GO:0009507 | P:formate catabolic process; F:formate dehydrogenase (NAD+) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding; C:mitochondrion; C:formate dehydrogenase complex; C:chloroplast | Acting on the CH-OH group of donors; Formate dehydrogenase; Acting on the aldehyde or oxo group of donors | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG645708.1 | Tinospora cordifolia | Transposon TX1 uncharacterized 149 kDa protein | 762 | P:GO:0006281; P:GO:0090305; F:GO:0003677; F:GO:0003824; F:GO:0004519; C:GO:0016020; C:GO:0016021 | P:DNA repair; P:nucleic acid phosphodiester bond hydrolysis; F:DNA binding; F:catalytic activity; F:endonuclease activity; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG649028.1 | Tinospora cordifolia | LIM domain-containing protein WLIM1-like | 549 | P:GO:0051017; F:GO:0046872; F:GO:0051015; C:GO:0005886; C:GO:0015629 | P:actin filament bundle assembly; F:metal ion binding; F:actin filament binding; C:plasma membrane; C:actin cytoskeleton | F:actin filament binding | view details | |||
| JG646359.1 | Tinospora cordifolia | Hypothetical predicted protein | 804 | no GO terms | view details | |||||
| JG650725.1 | Tinospora cordifolia | leucine aminopeptidase 1-like | 849 | P:GO:0006508; F:GO:0030145; F:GO:0070006; C:GO:0005737 | P:proteolysis; F:manganese ion binding; F:metalloaminopeptidase activity; C:cytoplasm | Acting on peptide bonds (peptidases) | P:proteolysis; P:protein metabolic process; F:manganese ion binding; F:metalloaminopeptidase activity; C:cytoplasm | view details | ||
| JG649413.1 | Tinospora cordifolia | salicylic acid-binding protein 2-like | 795 | P:GO:0009694; P:GO:0009696; F:GO:0080030; F:GO:0080031; F:GO:0080032 | P:jasmonic acid metabolic process; P:salicylic acid metabolic process; F:methyl indole-3-acetate esterase activity; F:methyl salicylate esterase activity; F:methyl jasmonate esterase activity | Carboxylesterase | no GO terms | view details | ||
| JG648975.1 | Tinospora cordifolia | putative P-loop containing nucleoside triphosphate hydrolase | 648 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| JG650442.1 | Tinospora cordifolia | salicylic acid-binding protein 2-like | 741 | P:GO:0009694; P:GO:0009696; F:GO:0016831; F:GO:0080030; F:GO:0080031; F:GO:0080032 | P:jasmonic acid metabolic process; P:salicylic acid metabolic process; F:carboxy-lyase activity; F:methyl indole-3-acetate esterase activity; F:methyl salicylate esterase activity; F:methyl jasmonate esterase activity | Carbon-carbon lyases; Carboxylesterase | no GO terms | view details | ||
| JG646913.1 | Tinospora cordifolia | SURNod19 domain-containing protein | 566 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650796.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 798 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG648763.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 830 | P:GO:0098542; F:GO:0016491; F:GO:0046872; C:GO:0016020 | P:defense response to other organism; F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648987.1 | Tinospora cordifolia | Negative regulator of systemic acquired resistance SNI1 | 496 | P:GO:0010113; P:GO:0016570; P:GO:0045892; F:GO:0000976; C:GO:0005634; C:GO:0016020 | P:negative regulation of systemic acquired resistance; P:histone modification; P:negative regulation of transcription, DNA-templated; F:transcription cis-regulatory region binding; C:nucleus; C:membrane | P:cellular response to DNA damage stimulus; P:negative regulation of defense response; P:negative regulation of transcription, DNA-templated; C:nucleus; C:Smc5-Smc6 complex | view details | |||
| JG645915.1 | Tinospora cordifolia | extradiol ring-cleavage dioxygenase | 844 | P:GO:0006725; F:GO:0008198; F:GO:0008270; F:GO:0016701; F:GO:0051213 | P:cellular aromatic compound metabolic process; F:ferrous iron binding; F:zinc ion binding; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen; F:dioxygenase activity | Acting on single donors with incorporation of molecular oxygen (oxygenases). The oxygen incorporated need not be derived from O(2) | P:cellular aromatic compound metabolic process; F:ferrous iron binding; F:zinc ion binding; F:oxidoreductase activity; F:oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | view details | ||
| JG649022.1 | Tinospora cordifolia | protein CREG1-like | 414 | C:GO:0005576; C:GO:0016021 | C:extracellular region; C:integral component of membrane | no GO terms | view details | |||
| JG646790.1 | Tinospora cordifolia | probable acyl-CoA dehydrogenase IBR3 | 829 | P:GO:0006468; P:GO:0006631; P:GO:0048767; F:GO:0004672; F:GO:0005524; F:GO:0016627; F:GO:0050660; C:GO:0005777 | P:protein phosphorylation; P:fatty acid metabolic process; P:root hair elongation; F:protein kinase activity; F:ATP binding; F:oxidoreductase activity, acting on the CH-CH group of donors; F:flavin adenine dinucleotide binding; C:peroxisome | Transferring phosphorus-containing groups; Acting on the CH-CH group of donors | no GO terms | view details | ||
| JG650128.1 | Tinospora cordifolia | methylenetetrahydrofolate reductase 2-like | 774 | P:GO:0006555; P:GO:0035999; F:GO:0106312; F:GO:0106313 | P:methionine metabolic process; P:tetrahydrofolate interconversion; F:methylenetetrahydrofolate reductase NADH activity; F:methylenetetrahydrofolate reductase NADPH activity | Methylenetetrahydrofolate reductase (NAD(P)H) | P:methionine metabolic process; F:methylenetetrahydrofolate reductase (NAD(P)H) activity | view details | ||
| JG647819.1 | Tinospora cordifolia | Myosin heavy chain, clone like | 783 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG646486.1 | Tinospora cordifolia | protein ZINC INDUCED FACILITATOR-LIKE 1-like | 805 | P:GO:0006817; P:GO:0050896; P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:phosphate ion transport; P:response to stimulus; P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity | view details | ||
| JG647924.1 | Tinospora cordifolia | auxin-responsive protein IAA1-like | 766 | P:GO:0006355; P:GO:0009734; C:GO:0005634 | P:regulation of transcription, DNA-templated; P:auxin-activated signaling pathway; C:nucleus | P:regulation of transcription, DNA-templated; F:protein binding; C:nucleus | view details | |||
| JG649177.1 | Tinospora cordifolia | Meiosis chromosome segregation family protein | 653 | no GO terms | view details | |||||
| JG648055.1 | Tinospora cordifolia | tubby-like F-box protein 8 | 554 | P:GO:0006355; P:GO:2000112 | P:regulation of transcription, DNA-templated; P:regulation of cellular macromolecule biosynthetic process | F:protein binding | view details | |||
| JG649961.1 | Tinospora cordifolia | R3H domain-containing protein 1 | 277 | F:GO:0003676 | F:nucleic acid binding | no GO terms | view details | |||
| JG650020.1 | Tinospora cordifolia | plant intracellular Ras-group-related LRR protein 5-like | 754 | no GO terms | view details | |||||
| JG646262.1 | Tinospora cordifolia | TPA_asm: hypothetical protein HUJ06_002193 | 692 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG648414.1 | Tinospora cordifolia | putative methyltransferase DDB_G0268948 | 830 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG645544.1 | Tinospora cordifolia | plant UBX domain-containing protein 8-like isoform X2 | 685 | no GO terms | view details | |||||
| JG646944.1 | Tinospora cordifolia | polyphenol oxidase I, chloroplastic-like | 735 | P:GO:0046148; P:GO:0090502; F:GO:0003676; F:GO:0004097; F:GO:0004503; F:GO:0004523; F:GO:0046872; C:GO:0016021 | P:pigment biosynthetic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:nucleic acid binding; F:catechol oxidase activity; F:tyrosinase activity; F:RNA-DNA hybrid ribonuclease activity; F:metal ion binding; C:integral component of membrane | Acting on ester bonds; Tyrosinase; Ribonuclease H; Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG650399.1 | Tinospora cordifolia | zinc finger protein ZAT10-like | 839 | P:GO:0006355; F:GO:0003700 | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity | F:DNA-binding transcription factor activity | view details | |||
| JG645656.1 | Tinospora cordifolia | Strictosidine synthase | 830 | P:GO:0009058; F:GO:0016844; C:GO:0005773; C:GO:0016021 | P:biosynthetic process; F:strictosidine synthase activity; C:vacuole; C:integral component of membrane | Strictosidine synthase | P:biosynthetic process; F:strictosidine synthase activity | view details | ||
| JG650257.1 | Tinospora cordifolia | leucine aminopeptidase 1-like | 673 | P:GO:0006508; F:GO:0030145; F:GO:0070006; C:GO:0005737 | P:proteolysis; F:manganese ion binding; F:metalloaminopeptidase activity; C:cytoplasm | Acting on peptide bonds (peptidases) | P:proteolysis; P:protein metabolic process; F:manganese ion binding; F:metalloaminopeptidase activity; C:cytoplasm | view details | ||
| JG646196.1 | Tinospora cordifolia | Hypothetical predicted protein | 724 | no GO terms | view details | |||||
| JG648706.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 814 | F:GO:0016491; F:GO:0046872; C:GO:0016020 | F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646128.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 853 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG650233.1 | Tinospora cordifolia | pyruvate kinase 1, cytosolic | 737 | P:GO:0006096; F:GO:0000287; F:GO:0004743; F:GO:0005524; F:GO:0016301; F:GO:0030955; C:GO:0005737 | P:glycolytic process; F:magnesium ion binding; F:pyruvate kinase activity; F:ATP binding; F:kinase activity; F:potassium ion binding; C:cytoplasm | Pyruvate kinase | P:glycolytic process; F:magnesium ion binding; F:catalytic activity; F:pyruvate kinase activity; F:potassium ion binding | view details | ||
| JG646347.1 | Tinospora cordifolia | 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial | 790 | P:GO:0007584; P:GO:0009083; F:GO:0003824; C:GO:0005947 | P:response to nutrient; P:branched-chain amino acid catabolic process; F:catalytic activity; C:mitochondrial alpha-ketoglutarate dehydrogenase complex | no GO terms | view details | |||
| JG650612.1 | Tinospora cordifolia | chaperone protein dnaJ A6-like | 774 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005783; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:endoplasmic reticulum; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG646003.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 814 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG648532.1 | Tinospora cordifolia | ap-1 complex subunit sigma-2 | 790 | no GO terms | view details | |||||
| JG645709.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 680 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646622.1 | Tinospora cordifolia | protein LUTEIN DEFICIENT 5, chloroplastic | 779 | P:GO:0016123; F:GO:0005506; F:GO:0010291; F:GO:0020037; F:GO:0070330 | P:xanthophyll biosynthetic process; F:iron ion binding; F:carotene beta-ring hydroxylase activity; F:heme binding; F:aromatase activity | Unspecific monooxygenase | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648415.1 | Tinospora cordifolia | transmembrane protein 33 homolog | 838 | P:GO:0016310; F:GO:0016301; C:GO:0016021 | P:phosphorylation; F:kinase activity; C:integral component of membrane | Transferring phosphorus-containing groups | C:integral component of membrane | view details | ||
| JG649527.1 | Tinospora cordifolia | probable 1-acylglycerol-3-phosphate O-acyltransferase | 776 | P:GO:0006654; P:GO:0055088; F:GO:0004623; F:GO:0042171 | P:phosphatidic acid biosynthetic process; P:lipid homeostasis; F:phospholipase A2 activity; F:lysophosphatidic acid acyltransferase activity | Carboxylesterase; Phospholipase A(2); Acyltransferases | P:proteolysis; F:peptidase activity | view details | ||
| JG645556.1 | Tinospora cordifolia | L-ascorbate oxidase homolog | 862 | F:GO:0005507; F:GO:0008447; C:GO:0016021 | F:copper ion binding; F:L-ascorbate oxidase activity; C:integral component of membrane | L-ascorbate oxidase | F:copper ion binding | view details | ||
| JG647651.1 | Tinospora cordifolia | Alpha/beta-Hydrolases superfamily protein | 765 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| JG648768.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 744 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG647445.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 742 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Acting on acid anhydrides; Lyases | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG646637.1 | Tinospora cordifolia | beta-glucosidase BoGH3B-like | 798 | P:GO:0009251; F:GO:0008422; C:GO:0005576 | P:glucan catabolic process; F:beta-glucosidase activity; C:extracellular region | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646787.1 | Tinospora cordifolia | 3,4-dihydroxy-2-butanone 4-phosphate synthase | 820 | P:GO:0009231; F:GO:0003935; F:GO:0005525; F:GO:0008686; F:GO:0046872; C:GO:0009507 | P:riboflavin biosynthetic process; F:GTP cyclohydrolase II activity; F:GTP binding; F:3,4-dihydroxy-2-butanone-4-phosphate synthase activity; F:metal ion binding; C:chloroplast | GTP cyclohydrolase II; 3,4-dihydroxy-2-butanone-4-phosphate synthase | P:riboflavin biosynthetic process; F:GTP cyclohydrolase II activity | view details | ||
| JG647840.1 | Tinospora cordifolia | polyphenol oxidase | 686 | P:GO:0046148; F:GO:0004097; F:GO:0008270 | P:pigment biosynthetic process; F:catechol oxidase activity; F:zinc ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG646860.1 | Tinospora cordifolia | putative indole-3-acetic acid-amido synthetase GH3.1 | 420 | no GO terms | view details | |||||
| JG650911.1 | Tinospora cordifolia | thaumatin-like protein | 777 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649652.1 | Tinospora cordifolia | hypothetical protein HHK36_027978 | 779 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG650960.1 | Tinospora cordifolia | uncharacterized MFS-type transporter YwfA-like isoform X2 | 809 | P:GO:0055085; F:GO:0022857; C:GO:0016021 | P:transmembrane transport; F:transmembrane transporter activity; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity | view details | ||
| JG647647.1 | Tinospora cordifolia | sulfate transporter 3.1-like | 722 | P:GO:1902358; F:GO:0008271; F:GO:0015293; F:GO:0015301; C:GO:0005887; C:GO:0009507 | P:sulfate transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:symporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane; C:chloroplast | Translocases | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:chloroplast; C:membrane | view details | ||
| JG647892.1 | Tinospora cordifolia | cystathionine gamma-synthase 1, chloroplastic-like | 854 | P:GO:0000096; P:GO:1901566; P:GO:1901605; F:GO:0003824 | P:sulfur amino acid metabolic process; P:organonitrogen compound biosynthetic process; P:alpha-amino acid metabolic process; F:catalytic activity | P:methionine biosynthetic process; P:transsulfuration; F:catalytic activity; F:cystathionine gamma-synthase activity; F:pyridoxal phosphate binding | view details | |||
| JG648162.1 | Tinospora cordifolia | putative methyltransferase DDB_G0268948 | 786 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG646276.1 | Tinospora cordifolia | beta-fructofuranosidase, insoluble isoenzyme CWINV1-like | 654 | P:GO:0005975; F:GO:0004564 | P:carbohydrate metabolic process; F:beta-fructofuranosidase activity | Beta-fructofuranosidase | no GO terms | view details | ||
| JG650672.1 | Tinospora cordifolia | 40S ribosomal protein S3a-like | 799 | P:GO:0006412; F:GO:0003735; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647621.1 | Tinospora cordifolia | acyl-coenzyme A oxidase 3, peroxisomal-like | 812 | P:GO:0033540; P:GO:0055088; F:GO:0003997; F:GO:0005504; F:GO:0008194; F:GO:0071949; C:GO:0005777 | P:fatty acid beta-oxidation using acyl-CoA oxidase; P:lipid homeostasis; F:acyl-CoA oxidase activity; F:fatty acid binding; F:UDP-glycosyltransferase activity; F:FAD binding; C:peroxisome | Glycosyltransferases; Acyl-CoA oxidase | P:fatty acid metabolic process; F:acyl-CoA oxidase activity; F:oxidoreductase activity, acting on the CH-CH group of donors; F:FAD binding; C:peroxisome | view details | ||
| JG650444.1 | Tinospora cordifolia | CAAX prenyl protease 1 homolog | 737 | P:GO:0071586; F:GO:0004222; F:GO:0046872; C:GO:0030176 | P:CAAX-box protein processing; F:metalloendopeptidase activity; F:metal ion binding; C:integral component of endoplasmic reticulum membrane | Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| JG648128.1 | Tinospora cordifolia | L-ascorbate oxidase-like | 798 | F:GO:0005507; F:GO:0008447; C:GO:0005576; C:GO:0016021 | F:copper ion binding; F:L-ascorbate oxidase activity; C:extracellular region; C:integral component of membrane | L-ascorbate oxidase | F:copper ion binding | view details | ||
| JG647253.1 | Tinospora cordifolia | protein NRT1/ PTR FAMILY 8.3 | 818 | P:GO:0006810; C:GO:0016020 | P:transport; C:membrane | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | |||
| JG648287.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase 14 | 734 | P:GO:0005975; F:GO:0042973; C:GO:0016021; C:GO:0046658 | P:carbohydrate metabolic process; F:glucan endo-1,3-beta-D-glucosidase activity; C:integral component of membrane; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646719.1 | Tinospora cordifolia | hypersensitive-induced reaction 1 protein | 765 | C:GO:0005737; C:GO:0043231 | C:cytoplasm; C:intracellular membrane-bounded organelle | no GO terms | view details | |||
| JG647022.1 | Tinospora cordifolia | unnamed protein product, partial | 718 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG649724.1 | Tinospora cordifolia | Tetratricopeptide TPR-1 | 777 | F:protein binding | view details | |||||
| JG650267.1 | Tinospora cordifolia | ruBisCO large subunit-binding protein subunit beta, chloroplastic | 722 | P:GO:0042026; F:GO:0005524; F:GO:0016887; C:GO:0005737 | P:protein refolding; F:ATP binding; F:ATP hydrolysis activity; C:cytoplasm | Acting on acid anhydrides | no IPS match | view details | ||
| JG647194.1 | Tinospora cordifolia | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | 838 | P:GO:0006413; P:GO:0030488; F:GO:0003743; F:GO:0008168; C:GO:0005634; C:GO:0031515 | P:translational initiation; P:tRNA methylation; F:translation initiation factor activity; F:methyltransferase activity; C:nucleus; C:tRNA (m1A) methyltransferase complex | Transferring one-carbon groups | P:tRNA methylation; C:tRNA (m1A) methyltransferase complex | view details | ||
| JG645533.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 741 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650231.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 798 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650662.1 | Tinospora cordifolia | Drug/metabolite transporter | 785 | P:GO:0015860; P:GO:1904823; F:GO:0005345; F:GO:0015211; C:GO:0016021 | P:purine nucleoside transmembrane transport; P:purine nucleobase transmembrane transport; F:purine nucleobase transmembrane transporter activity; F:purine nucleoside transmembrane transporter activity; C:integral component of membrane | Translocases | F:purine nucleoside transmembrane transporter activity; C:integral component of membrane | view details | ||
| JG647882.1 | Tinospora cordifolia | protein NO VEIN-like isoform X2 | 801 | no GO terms | view details | |||||
| JG645754.1 | Tinospora cordifolia | Diaminopimelate decarboxylase 2, chloroplastic | 764 | P:GO:0009089; F:GO:0008836 | P:lysine biosynthetic process via diaminopimelate; F:diaminopimelate decarboxylase activity | Diaminopimelate decarboxylase | F:catalytic activity | view details | ||
| JG646656.1 | Tinospora cordifolia | membrane steroid-binding protein 1 | 690 | C:GO:0005783; C:GO:0016021 | C:endoplasmic reticulum; C:integral component of membrane | no GO terms | view details | |||
| JG649791.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 778 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG645927.1 | Tinospora cordifolia | chaperone protein dnaJ A6-like | 807 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005783; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:endoplasmic reticulum; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG648745.1 | Tinospora cordifolia | putative methyltransferase DDB_G0268948 | 854 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG646366.1 | Tinospora cordifolia | mitochondrial phosphate carrier protein 3, mitochondrial-like | 872 | P:GO:1990547; F:GO:0005315; C:GO:0031305 | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity; C:integral component of mitochondrial inner membrane | Translocases | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity | view details | ||
| MS687928.1 | Tinospora cordifolia | FAD linked oxidase | 1770 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG648363.1 | Tinospora cordifolia | elongation factor 1-delta 1-like | 786 | P:GO:0006414; P:GO:0050790; F:GO:0003746; F:GO:0005085; C:GO:0005829; C:GO:0005853 | P:translational elongation; P:regulation of catalytic activity; F:translation elongation factor activity; F:guanyl-nucleotide exchange factor activity; C:cytosol; C:eukaryotic translation elongation factor 1 complex | P:translational elongation; F:translation elongation factor activity | view details | |||
| JG647415.1 | Tinospora cordifolia | probable glutathione S-transferase | 713 | P:GO:0006749; P:GO:0009636; F:GO:0004364; C:GO:0005829 | P:glutathione metabolic process; P:response to toxic substance; F:glutathione transferase activity; C:cytosol | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG648809.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein LOC107759405 | 392 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG649551.1 | Tinospora cordifolia | citrate synthase, mitochondrial | 806 | P:GO:0006101; F:GO:0004108 | P:citrate metabolic process; F:citrate (Si)-synthase activity | Citrate synthase (unknown stereospecificity) | F:acyltransferase activity, acyl groups converted into alkyl on transfer | view details | ||
| JG650074.1 | Tinospora cordifolia | THO complex subunit 2-like isoform X3 | 748 | P:GO:0006406; F:GO:0003729; C:GO:0000445 | P:mRNA export from nucleus; F:mRNA binding; C:THO complex part of transcription export complex | P:mRNA processing; P:mRNA export from nucleus; C:THO complex | view details | |||
| JG650272.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein LOC104607588 | 789 | P:GO:0007166 | P:cell surface receptor signaling pathway | F:protein binding | view details | |||
| JG647433.1 | Tinospora cordifolia | thaumatin-like protein | 771 | P:GO:0006952 | P:defense response | no GO terms | view details | |||
| JG650610.1 | Tinospora cordifolia | SUPPRESSOR OF ABI3-5 isoform X1 | 748 | P:GO:0000398; F:GO:0003723; C:GO:0005634 | P:mRNA splicing, via spliceosome; F:RNA binding; C:nucleus | no GO terms | view details | |||
| JG645664.1 | Tinospora cordifolia | UPF0481 protein At3g47200 | 820 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG645749.1 | Tinospora cordifolia | Glycoside hydrolase | 688 | P:GO:0009251; F:GO:0008422; F:GO:0102483; C:GO:0005576 | P:glucan catabolic process; F:beta-glucosidase activity; F:scopolin beta-glucosidase activity; C:extracellular region | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649993.1 | Tinospora cordifolia | receptor-like kinase TMK4 | 812 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:transmembrane receptor protein tyrosine kinase activity; F:protein binding | view details | ||
| JG648720.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 771 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG649403.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 808 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646480.1 | Tinospora cordifolia | Ketol-acid reductoisomerase, chloroplastic | 803 | P:GO:0009097; P:GO:0009099; F:GO:0004455; F:GO:0016853; F:GO:0046872; C:GO:0016021 | P:isoleucine biosynthetic process; P:valine biosynthetic process; F:ketol-acid reductoisomerase activity; F:isomerase activity; F:metal ion binding; C:integral component of membrane | Ketol-acid reductoisomerase (NADP(+)); Isomerases | P:branched-chain amino acid biosynthetic process; F:oxidoreductase activity | view details | ||
| JG648968.1 | Tinospora cordifolia | FAD-dependent urate hydroxylase-like | 644 | F:GO:0016491; C:GO:0016020 | F:oxidoreductase activity; C:membrane | Oxidoreductases | F:FAD binding | view details | ||
| JG645698.1 | Tinospora cordifolia | chaperone protein dnaJ A6-like | 795 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005783; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:endoplasmic reticulum; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG650949.1 | Tinospora cordifolia | ganglioside-induced differentiation-associated protein 2 | 804 | no GO terms | view details | |||||
| JG649522.1 | Tinospora cordifolia | L-ascorbate oxidase homolog | 879 | F:GO:0005507; F:GO:0008447; C:GO:0016021 | F:copper ion binding; F:L-ascorbate oxidase activity; C:integral component of membrane | L-ascorbate oxidase | F:copper ion binding | view details | ||
| JG650682.1 | Tinospora cordifolia | pathogen-related protein-like | 719 | no GO terms | view details | |||||
| JG646374.1 | Tinospora cordifolia | hypersensitive-induced reaction 1 protein | 797 | C:GO:0005737; C:GO:0043231 | C:cytoplasm; C:intracellular membrane-bounded organelle | no GO terms | view details | |||
| JG647975.1 | Tinospora cordifolia | DUF1336 domain-containing protein | 746 | no GO terms | view details | |||||
| JG648824.1 | Tinospora cordifolia | desiccation-related protein At2g46140 | 386 | P:GO:0009269 | P:response to desiccation | no GO terms | view details | |||
| JG646315.1 | Tinospora cordifolia | Stress up-regulated Nod 19 protein | 783 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| LQ667239.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 960 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | view details | ||
| JG648557.1 | Tinospora cordifolia | SKP1-like protein 1B | 538 | P:GO:0016310; P:GO:0016567; P:GO:0031146; F:GO:0016301; F:GO:0097602; C:GO:0005634; C:GO:0005737 | P:phosphorylation; P:protein ubiquitination; P:SCF-dependent proteasomal ubiquitin-dependent protein catabolic process; F:kinase activity; F:cullin family protein binding; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | P:ubiquitin-dependent protein catabolic process | view details | ||
| JG648559.1 | Tinospora cordifolia | protein GRAVITROPIC IN THE LIGHT 1-like | 813 | P:GO:0009639; P:GO:0009959 | P:response to red or far red light; P:negative gravitropism | P:response to red or far red light; P:negative gravitropism | view details | |||
| JG649320.1 | Tinospora cordifolia | Non-symbiotic hemoglobin | 730 | P:GO:0015671; F:GO:0005344; F:GO:0019825; F:GO:0020037; F:GO:0046872 | P:oxygen transport; F:oxygen carrier activity; F:oxygen binding; F:heme binding; F:metal ion binding | F:oxygen binding; F:heme binding | view details | |||
| JG649873.1 | Tinospora cordifolia | 40S ribosomal protein S19-1-like | 538 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; F:GO:0016740; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; F:transferase activity; C:cytosolic small ribosomal subunit | Transferases | P:translation; F:structural constituent of ribosome; C:ribosome | view details | ||
| JG646616.1 | Tinospora cordifolia | selenium-binding protein 1 | 764 | F:GO:0008430; F:GO:0018549 | F:selenium binding; F:methanethiol oxidase activity | Methanethiol oxidase | F:selenium binding | view details | ||
| JG650764.1 | Tinospora cordifolia | JmjC domain | 853 | P:GO:0034720; P:GO:0070544; F:GO:0032453; F:GO:0046872; F:GO:0051864; C:GO:0005730 | P:histone H3-K4 demethylation; P:histone H3-K36 demethylation; F:histone H3-methyl-lysine-4 demethylase activity; F:metal ion binding; F:histone H3-methyl-lysine-36 demethylase activity; C:nucleolus | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no GO terms | view details | ||
| JG646679.1 | Tinospora cordifolia | protein ASPARTIC PROTEASE IN GUARD CELL 1 | 826 | P:GO:0006508; F:GO:0004190; C:GO:0016021 | P:proteolysis; F:aspartic-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG646273.1 | Tinospora cordifolia | Bromo adjacent homology (BAH) domain | 788 | P:GO:0006351; F:GO:0003682 | P:transcription, DNA-templated; F:chromatin binding | F:chromatin binding | view details | |||
| JG650937.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 802 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG648817.1 | Tinospora cordifolia | Thaumatin-like protein | 512 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG646765.1 | Tinospora cordifolia | gamma carbonic anhydrase-like 2, mitochondrial | 825 | no GO terms | view details | |||||
| JG649398.1 | Tinospora cordifolia | Stress up-regulated Nod 19 protein | 773 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650067.1 | Tinospora cordifolia | armadillo repeat-containing protein 8 | 733 | P:GO:0043161; C:GO:0005634; C:GO:0005737; C:GO:0034657 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; C:nucleus; C:cytoplasm; C:GID complex | no GO terms | view details | |||
| JG648893.1 | Tinospora cordifolia | leucine aminopeptidase 1-like | 840 | P:GO:0006508; F:GO:0030145; F:GO:0070006; C:GO:0005737 | P:proteolysis; F:manganese ion binding; F:metalloaminopeptidase activity; C:cytoplasm | Acting on peptide bonds (peptidases) | P:proteolysis; P:protein metabolic process; F:manganese ion binding; F:metalloaminopeptidase activity; C:cytoplasm | view details | ||
| JG651011.1 | Tinospora cordifolia | chaperone protein dnaJ A6-like | 723 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005783; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:endoplasmic reticulum; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG646098.1 | Tinospora cordifolia | probable WRKY transcription factor 17 | 821 | P:GO:0006355; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG650768.1 | Tinospora cordifolia | Inhibitor of Bruton tyrosine kinase | 814 | P:GO:0016310; F:GO:0016301 | P:phosphorylation; F:kinase activity | no GO terms | view details | |||
| JG648780.1 | Tinospora cordifolia | phosphoglucomutase, cytoplasmic | 256 | P:GO:0006006; F:GO:0000287; F:GO:0004614; C:GO:0005829 | P:glucose metabolic process; F:magnesium ion binding; F:phosphoglucomutase activity; C:cytosol | Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | P:carbohydrate metabolic process; F:phosphoglucomutase activity; F:intramolecular transferase activity, phosphotransferases | view details | ||
| JG645525.1 | Tinospora cordifolia | ATP sulfurylase 1, chloroplastic | 679 | P:GO:0000103; P:GO:0016310; F:GO:0004020; F:GO:0004781; F:GO:0005524 | P:sulfate assimilation; P:phosphorylation; F:adenylylsulfate kinase activity; F:sulfate adenylyltransferase (ATP) activity; F:ATP binding | Sulfate adenylyltransferase; Adenylyl-sulfate kinase | no GO terms | view details | ||
| JG647562.1 | Tinospora cordifolia | TRAF-like family protein | 736 | P:GO:0009058; F:GO:0016787; F:GO:0030170 | P:biosynthetic process; F:hydrolase activity; F:pyridoxal phosphate binding | Hydrolases | no GO terms | view details | ||
| JG649235.1 | Tinospora cordifolia | Stress up-regulated Nod 19 protein | 74 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no IPS match | view details | |||
| JG647594.1 | Tinospora cordifolia | putative methyltransferase DDB_G0268948 | 835 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG645764.1 | Tinospora cordifolia | Inositol-1,4,5-trisphosphate 5-phosphatase 4 isoform 2 | 783 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG648328.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein LOC104609646 | 813 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG648623.1 | Tinospora cordifolia | glutathione reductase, cytosolic | 756 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG649094.1 | Tinospora cordifolia | pathogen-related protein-like | 661 | no GO terms | view details | |||||
| JG646497.1 | Tinospora cordifolia | putative G3BP-like protein | 560 | F:GO:0003729; C:GO:0005829; C:GO:1990904 | F:mRNA binding; C:cytosol; C:ribonucleoprotein complex | F:nucleic acid binding; F:RNA binding | view details | |||
| JG648425.1 | Tinospora cordifolia | UPF0415 protein C7orf25 homolog | 813 | no GO terms | view details | |||||
| JG648792.1 | Tinospora cordifolia | L-ascorbate oxidase homolog | 493 | F:GO:0005507; F:GO:0016491 | F:copper ion binding; F:oxidoreductase activity | Oxidoreductases | F:copper ion binding | view details | ||
| JG650342.1 | Tinospora cordifolia | probable N-succinyldiaminopimelate aminotransferase DapC | 806 | P:GO:0009058; P:GO:0097052; F:GO:0016212; F:GO:0030170; C:GO:0005737 | P:biosynthetic process; P:L-kynurenine metabolic process; F:kynurenine-oxoglutarate transaminase activity; F:pyridoxal phosphate binding; C:cytoplasm | Kynurenine--oxoglutarate transaminase | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG646932.1 | Tinospora cordifolia | katanin p60 ATPase-containing subunit A1 | 203 | P:GO:0030433; P:GO:0045899; P:GO:0051301; P:GO:1901800; F:GO:0005524; F:GO:0016887; F:GO:0036402; C:GO:0008540; C:GO:0016021; C:GO:0031597 | P:ubiquitin-dependent ERAD pathway; P:positive regulation of RNA polymerase II transcription preinitiation complex assembly; P:cell division; P:positive regulation of proteasomal protein catabolic process; F:ATP binding; F:ATP hydrolysis activity; F:proteasome-activating activity; C:proteasome regulatory particle, base subcomplex; C:integral component of membrane; C:cytosolic proteasome complex | Acting on acid anhydrides; Proteasome ATPase | no IPS match | view details | ||
| JG647144.1 | Tinospora cordifolia | EEF1A lysine methyltransferase 2-like | 749 | P:GO:0018026; P:GO:0018027; F:GO:0016279; C:GO:0005737 | P:peptidyl-lysine monomethylation; P:peptidyl-lysine dimethylation; F:protein-lysine N-methyltransferase activity; C:cytoplasm | Transferring one-carbon groups | no GO terms | view details | ||
| JG646646.1 | Tinospora cordifolia | 60S ribosomal protein L6 | 785 | P:GO:0000027; P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:ribosomal large subunit assembly; P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647379.1 | Tinospora cordifolia | plant UBX domain-containing protein 7 isoform X2 | 550 | P:GO:0043161; F:GO:0043130; C:GO:0005634 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; F:ubiquitin binding; C:nucleus | F:protein binding | view details | |||
| JG645537.1 | Tinospora cordifolia | succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial-like | 800 | P:GO:0006099; P:GO:0006121; F:GO:0008177; F:GO:0009055; F:GO:0050660; C:GO:0005749 | P:tricarboxylic acid cycle; P:mitochondrial electron transport, succinate to ubiquinone; F:succinate dehydrogenase (ubiquinone) activity; F:electron transfer activity; F:flavin adenine dinucleotide binding; C:mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | Succinate dehydrogenase (quinone) | no GO terms | view details | ||
| JG650378.1 | Tinospora cordifolia | acyl-coenzyme A oxidase 4, peroxisomal | 809 | P:GO:0006635; F:GO:0003995; F:GO:0050660; C:GO:0005777; C:GO:0016021 | P:fatty acid beta-oxidation; F:acyl-CoA dehydrogenase activity; F:flavin adenine dinucleotide binding; C:peroxisome; C:integral component of membrane | Acting on the CH-CH group of donors | P:fatty acid beta-oxidation; F:acyl-CoA dehydrogenase activity; F:oxidoreductase activity, acting on the CH-CH group of donors; F:flavin adenine dinucleotide binding | view details | ||
| JG649916.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 517 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648533.1 | Tinospora cordifolia | cystathionine gamma-synthase 1, chloroplastic-like | 831 | P:GO:0000096; P:GO:1901566; P:GO:1901605; F:GO:0003824 | P:sulfur amino acid metabolic process; P:organonitrogen compound biosynthetic process; P:alpha-amino acid metabolic process; F:catalytic activity | P:methionine biosynthetic process; P:transsulfuration; F:catalytic activity; F:cystathionine gamma-synthase activity; F:pyridoxal phosphate binding | view details | |||
| JG650673.1 | Tinospora cordifolia | BEL1-like homeodomain protein 1 | 785 | P:GO:0006355; F:GO:0003677 | P:regulation of transcription, DNA-templated; F:DNA binding | no GO terms | view details | |||
| JG648579.1 | Tinospora cordifolia | non-symbiotic hemoglobin 2 | 792 | P:GO:0015671; F:GO:0005344; F:GO:0019825; F:GO:0020037; F:GO:0046872 | P:oxygen transport; F:oxygen carrier activity; F:oxygen binding; F:heme binding; F:metal ion binding | F:oxygen binding; F:heme binding | view details | |||
| JG646519.1 | Tinospora cordifolia | Nhl domain-containing protein | 813 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650813.1 | Tinospora cordifolia | Bromo adjacent homology (BAH) domain | 815 | F:GO:0003682; C:GO:0005634 | F:chromatin binding; C:nucleus | no GO terms | view details | |||
| JG648103.1 | Tinospora cordifolia | bark storage protein A-like | 770 | P:GO:0009116; F:GO:0003824 | P:nucleoside metabolic process; F:catalytic activity | P:nucleoside metabolic process; F:catalytic activity | view details | |||
| JG647037.1 | Tinospora cordifolia | protein SMG7-like | 782 | P:GO:0000184; F:GO:0042162; F:GO:0070034; C:GO:0005697 | P:nuclear-transcribed mRNA catabolic process, nonsense-mediated decay; F:telomeric DNA binding; F:telomerase RNA binding; C:telomerase holoenzyme complex | F:protein binding | view details | |||
| JG649128.1 | Tinospora cordifolia | ninja-family protein mc410 | 876 | P:GO:0007165 | P:signal transduction | P:signal transduction | view details | |||
| JG647547.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 790 | no GO terms | view details | |||||
| JG649601.1 | Tinospora cordifolia | putative actin cross-linking protein | 727 | no GO terms | view details | |||||
| JG645746.1 | Tinospora cordifolia | FAD linked oxidase | 821 | F:GO:0050328; F:GO:0071949 | F:tetrahydroberberine oxidase activity; F:FAD binding | Tetrahydroberberine oxidase | F:oxidoreductase activity; F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| JG649404.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 743 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG648918.1 | Tinospora cordifolia | phosphatidylinositol transfer protein 3-like | 798 | P:GO:0015914; P:GO:0120009; F:GO:0008526; C:GO:0016020 | P:phospholipid transport; P:intermembrane lipid transfer; F:phosphatidylinositol transfer activity; C:membrane | Translocases | no GO terms | view details | ||
| JG647023.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 807 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646866.1 | Tinospora cordifolia | peptide methionine sulfoxide reductase B5-like | 536 | P:GO:0006979; P:GO:0030091; F:GO:0033743; F:GO:0046872 | P:response to oxidative stress; P:protein repair; F:peptide-methionine (R)-S-oxide reductase activity; F:metal ion binding | Peptide-methionine (R)-S-oxide reductase | P:response to oxidative stress; P:protein repair; F:oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor; F:peptide-methionine (R)-S-oxide reductase activity | view details | ||
| JG649667.1 | Tinospora cordifolia | glutathione S-transferase U17-like | 765 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity; F:protein binding | view details | ||
| JG646449.1 | Tinospora cordifolia | hypersensitive-induced reaction 1 protein | 790 | C:GO:0005737; C:GO:0043231 | C:cytoplasm; C:intracellular membrane-bounded organelle | no GO terms | view details | |||
| JG647422.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase 14-like | 694 | P:GO:0005975; F:GO:0042973; C:GO:0016021; C:GO:0046658 | P:carbohydrate metabolic process; F:glucan endo-1,3-beta-D-glucosidase activity; C:integral component of membrane; C:anchored component of plasma membrane | Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645639.1 | Tinospora cordifolia | pectinesterase 2 | 792 | P:GO:0042545; P:GO:0043086; P:GO:0045490; F:GO:0030599; F:GO:0045330; F:GO:0046910; C:GO:0005576 | P:cell wall modification; P:negative regulation of catalytic activity; P:pectin catabolic process; F:pectinesterase activity; F:aspartyl esterase activity; F:pectinesterase inhibitor activity; C:extracellular region | Pectinesterase | F:enzyme inhibitor activity | view details | ||
| JG646864.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 626 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG650239.1 | Tinospora cordifolia | Non-symbiotic hemoglobin | 808 | P:GO:0015671; F:GO:0005344; F:GO:0019825; F:GO:0020037; F:GO:0046872 | P:oxygen transport; F:oxygen carrier activity; F:oxygen binding; F:heme binding; F:metal ion binding | F:oxygen binding; F:heme binding | view details | |||
| JG650131.1 | Tinospora cordifolia | IST1 homolog | 814 | P:GO:0015031 | P:protein transport | P:protein transport | view details | |||
| JG650637.1 | Tinospora cordifolia | 40S ribosomal protein S3a | 827 | P:GO:0006412; F:GO:0003735; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648326.1 | Tinospora cordifolia | putative methyltransferase DDB_G0268948 | 818 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG647907.1 | Tinospora cordifolia | cleavage stimulating factor 64-like isoform X1 | 811 | P:GO:0016070; F:GO:0003676 | P:RNA metabolic process; F:nucleic acid binding | no GO terms | view details | |||
| JG648089.1 | Tinospora cordifolia | Armadillo repeat only 2 | 746 | P:GO:0007166 | P:cell surface receptor signaling pathway | P:cell surface receptor signaling pathway | view details | |||
| JG647814.1 | Tinospora cordifolia | desiccation-related protein At2g46140 | 801 | P:GO:0009269; C:GO:0016021 | P:response to desiccation; C:integral component of membrane | no GO terms | view details | |||
| JG648010.1 | Tinospora cordifolia | heterogeneous nuclear ribonucleoprotein 1 | 722 | F:GO:0003723; C:GO:0016021 | F:RNA binding; C:integral component of membrane | F:nucleic acid binding; F:RNA binding | view details | |||
| JG650255.1 | Tinospora cordifolia | puromycin-sensitive aminopeptidase-like isoform X1 | 809 | P:GO:0006508; F:GO:0004177; F:GO:0008237; F:GO:0008270; C:GO:0016021 | P:proteolysis; F:aminopeptidase activity; F:metallopeptidase activity; F:zinc ion binding; C:integral component of membrane | Acting on peptide bonds (peptidases) | F:zinc ion binding | view details | ||
| JG649612.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 652 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Acting on acid anhydrides; Lyases | no IPS match | view details | ||
| JG647582.1 | Tinospora cordifolia | O-glucosyltransferase rumi homolog | 804 | F:GO:0016740; C:GO:0016021 | F:transferase activity; C:integral component of membrane | Transferases | no GO terms | view details | ||
| JG650111.1 | Tinospora cordifolia | plant intracellular Ras-group-related LRR protein 5-like | 823 | no GO terms | view details | |||||
| JG650797.1 | Tinospora cordifolia | 14-3-3-like protein | 812 | P:GO:0007165; C:GO:0005737 | P:signal transduction; C:cytoplasm | no GO terms | view details | |||
| JG650092.1 | Tinospora cordifolia | chaperone protein dnaJ A6-like | 722 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005783; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:endoplasmic reticulum; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG650421.1 | Tinospora cordifolia | xanthotoxin 5-hydroxylase CYP82C4-like | 798 | F:GO:0016491; F:GO:0046872; C:GO:0016020 | F:oxidoreductase activity; F:metal ion binding; C:membrane | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645546.1 | Tinospora cordifolia | sulfate transporter 1.3-like | 718 | P:GO:0090502; P:GO:1902358; F:GO:0003676; F:GO:0004523; F:GO:0008271; F:GO:0015301; C:GO:0005887 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; P:sulfate transmembrane transport; F:nucleic acid binding; F:RNA-DNA hybrid ribonuclease activity; F:secondary active sulfate transmembrane transporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane | Acting on ester bonds; Translocases; Ribonuclease H | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG649075.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 725 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646440.1 | Tinospora cordifolia | steroid 5-alpha-reductase DET2 isoform X2 | 845 | P:GO:0006629; F:GO:0016627; C:GO:0016021 | P:lipid metabolic process; F:oxidoreductase activity, acting on the CH-CH group of donors; C:integral component of membrane | Acting on the CH-CH group of donors | P:lipid metabolic process; F:oxidoreductase activity, acting on the CH-CH group of donors | view details | ||
| JG650259.1 | Tinospora cordifolia | putative leucine-rich repeat-containing protein DDB_G0290503 isoform X1 | 747 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG649249.1 | Tinospora cordifolia | Formin-like protein 18 | 766 | no GO terms | view details | |||||
| JG649185.1 | Tinospora cordifolia | putative ribonuclease H protein | 829 | P:GO:0006281; P:GO:0015074; P:GO:0090305; F:GO:0003676; F:GO:0003677; F:GO:0003824; F:GO:0004519 | P:DNA repair; P:DNA integration; P:nucleic acid phosphodiester bond hydrolysis; F:nucleic acid binding; F:DNA binding; F:catalytic activity; F:endonuclease activity | no GO terms | view details | |||
| JG646843.1 | Tinospora cordifolia | phospholipase A1-Igamma1, chloroplastic-like | 658 | P:GO:0006629 | P:lipid metabolic process | no GO terms | view details | |||
| JG650149.1 | Tinospora cordifolia | Xanthotoxin 5-hydroxylase CYP82C4 | 704 | F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650001.1 | Tinospora cordifolia | chaperone protein dnaJ A6-like | 761 | P:GO:0009408; P:GO:0042026; F:GO:0005524; F:GO:0030544; F:GO:0046872; F:GO:0051082; F:GO:0051087; C:GO:0005783; C:GO:0005829 | P:response to heat; P:protein refolding; F:ATP binding; F:Hsp70 protein binding; F:metal ion binding; F:unfolded protein binding; F:chaperone binding; C:endoplasmic reticulum; C:cytosol | P:protein folding; F:Hsp70 protein binding; F:heat shock protein binding; F:unfolded protein binding | view details | |||
| JG650514.1 | Tinospora cordifolia | probable WRKY transcription factor 31 | 815 | P:GO:0006355; F:GO:0003677; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | F:DNA-binding transcription factor activity | view details | |||
| JG648309.1 | Tinospora cordifolia | glycosyl hydrolase family protein | 774 | P:GO:0005975; F:GO:0004553; C:GO:0005576 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; C:extracellular region | Glycosylases | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646413.1 | Tinospora cordifolia | phospholipase A1-Igamma1, chloroplastic-like | 861 | P:GO:0006629 | P:lipid metabolic process | P:lipid metabolic process | view details | |||
| JG649300.1 | Tinospora cordifolia | Bromo adjacent homology (BAH) domain | 713 | F:GO:0003682; C:GO:0005634 | F:chromatin binding; C:nucleus | no GO terms | view details | |||
| JG648236.1 | Tinospora cordifolia | transmembrane protein 205 | 537 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG649900.1 | Tinospora cordifolia | Transmembrane protein 70, mitochondrial | 426 | P:GO:0033615; F:GO:0003677; C:GO:0032592 | P:mitochondrial proton-transporting ATP synthase complex assembly; F:DNA binding; C:integral component of mitochondrial membrane | no GO terms | view details | |||
| JG649554.1 | Tinospora cordifolia | 40S ribosomal protein S3a-like | 785 | P:GO:0006412; F:GO:0003735; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648175.1 | Tinospora cordifolia | protein SCO1 homolog 1, mitochondrial | 822 | P:GO:0033617; C:GO:0005739; C:GO:0016021 | P:mitochondrial cytochrome c oxidase assembly; C:mitochondrion; C:integral component of membrane | no GO terms | view details | |||
| JG647577.1 | Tinospora cordifolia | Stress up-regulated Nod 19 | 836 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646349.1 | Tinospora cordifolia | mediator of RNA polymerase II transcription subunit 13 isoform X1 | 681 | C:GO:0005634 | C:nucleus | no GO terms | view details | |||
| JG647441.1 | Tinospora cordifolia | calmodulin-binding protein 25-like | 521 | P:GO:0006970; F:GO:0005516; C:GO:0005634 | P:response to osmotic stress; F:calmodulin binding; C:nucleus | no GO terms | view details | |||
| JG650174.1 | Tinospora cordifolia | polyphenol oxidase I, chloroplastic-like | 480 | P:GO:0046148; P:GO:0090502; F:GO:0003676; F:GO:0004097; F:GO:0004523; F:GO:0046872; C:GO:0016021 | P:pigment biosynthetic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:nucleic acid binding; F:catechol oxidase activity; F:RNA-DNA hybrid ribonuclease activity; F:metal ion binding; C:integral component of membrane | Acting on ester bonds; Ribonuclease H; Catechol oxidase | no GO terms | view details | ||
| JG649163.1 | Tinospora cordifolia | serine/threonine-protein kinase CTR1 isoform X1 | 761 | P:GO:0006073; P:GO:0016310; F:GO:0000166; F:GO:0004672 | P:cellular glucan metabolic process; P:phosphorylation; F:nucleotide binding; F:protein kinase activity | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG649073.1 | Tinospora cordifolia | ethylene-responsive transcription factor CRF4-like | 237 | P:GO:0006355; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of transcription, DNA-templated; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | no IPS match | view details | |||
| JG649089.1 | Tinospora cordifolia | formate dehydrogenase, mitochondrial | 705 | P:GO:0042183; F:GO:0008863; F:GO:0016616; F:GO:0051287; C:GO:0005739; C:GO:0009326; C:GO:0009507 | P:formate catabolic process; F:formate dehydrogenase (NAD+) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding; C:mitochondrion; C:formate dehydrogenase complex; C:chloroplast | Acting on the CH-OH group of donors; Formate dehydrogenase; Acting on the aldehyde or oxo group of donors | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG650935.1 | Tinospora cordifolia | Meiosis chromosome segregation family protein | 808 | no GO terms | view details | |||||
| JG647263.1 | Tinospora cordifolia | ankyrin repeat domain-containing protein 13C-B | 814 | C:GO:0005737; C:GO:0016020 | C:cytoplasm; C:membrane | no GO terms | view details | |||
| JG647917.1 | Tinospora cordifolia | beta-glucosidase BoGH3B-like | 728 | P:GO:0009251; F:GO:0008422; C:GO:0005576; C:GO:0016021 | P:glucan catabolic process; F:beta-glucosidase activity; C:extracellular region; C:integral component of membrane | Beta-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649936.1 | Tinospora cordifolia | cytochrome P450 94B3 | 401 | P:GO:0006414; P:GO:0006508; F:GO:0003746; F:GO:0004497; F:GO:0005506; F:GO:0008234; F:GO:0016705; F:GO:0020037; C:GO:0005853 | P:translational elongation; P:proteolysis; F:translation elongation factor activity; F:monooxygenase activity; F:iron ion binding; F:cysteine-type peptidase activity; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:eukaryotic translation elongation factor 1 complex | Acting on peptide bonds (peptidases); Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650546.1 | Tinospora cordifolia | IRK-interacting protein | 816 | P:GO:0009639; P:GO:0009959 | P:response to red or far red light; P:negative gravitropism | P:response to red or far red light; P:negative gravitropism | view details | |||
| JG648020.1 | Tinospora cordifolia | ARM REPEAT PROTEIN INTERACTING WITH ABF2-like | 696 | P:GO:0007166 | P:cell surface receptor signaling pathway | P:cell surface receptor signaling pathway; F:protein binding | view details | |||
| JG648059.1 | Tinospora cordifolia | caffeic acid O-methyltransferase | 805 | P:GO:0019438; F:GO:0008171 | P:aromatic compound biosynthetic process; F:O-methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity; F:O-methyltransferase activity; F:protein dimerization activity | view details | ||
| JG647365.1 | Tinospora cordifolia | protein translocase subunit SECA2, chloroplastic isoform X4 | 715 | P:GO:0008152; P:GO:0015031; F:GO:0005488; F:GO:0016740 | P:metabolic process; P:protein transport; F:binding; F:transferase activity | Transferases | no GO terms | view details | ||
| JG650431.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 715 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG648801.1 | Tinospora cordifolia | protoporphyrinogen oxidase 1, chloroplastic | 801 | P:GO:0006782; F:GO:0004729; C:GO:0009507 | P:protoporphyrinogen IX biosynthetic process; F:oxygen-dependent protoporphyrinogen oxidase activity; C:chloroplast | Protoporphyrinogen oxidase | P:porphyrin-containing compound biosynthetic process; F:oxygen-dependent protoporphyrinogen oxidase activity; F:oxidoreductase activity | view details | ||
| JG650220.1 | Tinospora cordifolia | protein SUPPRESSOR OF GENE SILENCING 3 | 691 | P:GO:0031047; P:GO:0051607 | P:gene silencing by RNA; P:defense response to virus | P:gene silencing by RNA; P:defense response to virus | view details | |||
| JG650404.1 | Tinospora cordifolia | UDP-glucose 6-dehydrogenase family protein | 855 | P:GO:0000271; P:GO:0006065; F:GO:0003979; F:GO:0051287 | P:polysaccharide biosynthetic process; P:UDP-glucuronate biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:NAD binding | UDP-glucose 6-dehydrogenase | P:polysaccharide biosynthetic process; F:UDP-glucose 6-dehydrogenase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG649750.1 | Tinospora cordifolia | la-related protein 1A | 641 | F:GO:0003723; F:GO:0008168 | F:RNA binding; F:methyltransferase activity | Transferring one-carbon groups | no GO terms | view details | ||
| JG649140.1 | Tinospora cordifolia | Plasma membrane isoform 1 | 292 | C:GO:0005840 | C:ribosome | no GO terms | view details | |||
| JG648663.1 | Tinospora cordifolia | non-functional NADPH-dependent codeinone reductase 2-like | 714 | F:GO:0004032; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; C:cytosol | Aldose reductase | F:oxidoreductase activity | view details | ||
| JG649428.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 812 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG646312.1 | Tinospora cordifolia | sulfate transporter 1.3-like | 736 | P:GO:0090502; P:GO:1902358; F:GO:0003676; F:GO:0004523; F:GO:0008271; F:GO:0015301; C:GO:0005887 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; P:sulfate transmembrane transport; F:nucleic acid binding; F:RNA-DNA hybrid ribonuclease activity; F:secondary active sulfate transmembrane transporter activity; F:anion:anion antiporter activity; C:integral component of plasma membrane | Acting on ester bonds; Translocases; Ribonuclease H | P:sulfate transport; P:transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:sulfate transmembrane transporter activity; C:membrane; C:integral component of membrane | view details | ||
| JG648736.1 | Tinospora cordifolia | 40S ribosomal protein S2-4-like | 770 | P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0005829; C:GO:0015935 | P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosol; C:small ribosomal subunit | P:translation; F:RNA binding; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG645720.1 | Tinospora cordifolia | Bromo adjacent homology (BAH) domain | 786 | F:GO:0003682; C:GO:0005634 | F:chromatin binding; C:nucleus | no GO terms | view details | |||
| JG650242.1 | Tinospora cordifolia | desiccation-related protein At2g46140 | 665 | P:GO:0009269; C:GO:0016021 | P:response to desiccation; C:integral component of membrane | no GO terms | view details | |||
| JG648357.1 | Tinospora cordifolia | WD40 repeat | 855 | P:GO:0006914; C:GO:0043231 | P:autophagy; C:intracellular membrane-bounded organelle | F:protein binding | view details | |||
| JG649142.1 | Tinospora cordifolia | 14-3-3-like protein | 409 | P:GO:0007165; C:GO:0005737 | P:signal transduction; C:cytoplasm | no GO terms | view details | |||
| JG648226.1 | Tinospora cordifolia | egalitarian protein homolog | 760 | P:GO:0090305; F:GO:0003723; F:GO:0008408 | P:nucleic acid phosphodiester bond hydrolysis; F:RNA binding; F:3 -5 exonuclease activity | Acting on ester bonds | P:nucleobase-containing compound metabolic process; F:nucleic acid binding; F:3 -5 exonuclease activity | view details | ||
| JG647216.1 | Tinospora cordifolia | adenylosuccinate synthetase 2, chloroplastic | 810 | P:GO:0044208; P:GO:0046040; F:GO:0000287; F:GO:0004019; F:GO:0005525; C:GO:0009507 | P:de novo AMP biosynthetic process; P:IMP metabolic process; F:magnesium ion binding; F:adenylosuccinate synthase activity; F:GTP binding; C:chloroplast | Adenylosuccinate synthase | P:purine nucleotide biosynthetic process; F:adenylosuccinate synthase activity; F:GTP binding | view details | ||
| JG645966.1 | Tinospora cordifolia | serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B beta isoform-like | 683 | P:GO:0007165; P:GO:0050790; F:GO:0019888; C:GO:0000159 | P:signal transduction; P:regulation of catalytic activity; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex | P:signal transduction; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex | view details | |||
| JG645730.1 | Tinospora cordifolia | 5-methylthioadenosine/S-adenosylhomocysteine nucleosidase 2 | 724 | P:GO:0009116; P:GO:0019509; F:GO:0008930 | P:nucleoside metabolic process; P:L-methionine salvage from methylthioadenosine; F:methylthioadenosine nucleosidase activity | Methylthioadenosine nucleosidase | P:nucleoside metabolic process; P:L-methionine salvage from methylthioadenosine; F:catalytic activity; F:methylthioadenosine nucleosidase activity | view details | ||
| JG646675.1 | Tinospora cordifolia | adenylosuccinate synthetase 2, chloroplastic | 736 | P:GO:0044208; P:GO:0046040; F:GO:0000287; F:GO:0004019; F:GO:0005525; C:GO:0009507 | P:de novo AMP biosynthetic process; P:IMP metabolic process; F:magnesium ion binding; F:adenylosuccinate synthase activity; F:GTP binding; C:chloroplast | Adenylosuccinate synthase | P:purine nucleotide biosynthetic process; F:adenylosuccinate synthase activity; F:GTP binding | view details | ||
| JG647239.1 | Tinospora cordifolia | DNA replication licensing factor MCM6 | 764 | P:GO:0000727; P:GO:0006268; P:GO:0006270; P:GO:1902969; F:GO:0003697; F:GO:0005524; F:GO:0016887; F:GO:1990518; C:GO:0000347; C:GO:0042555 | P:double-strand break repair via break-induced replication; P:DNA unwinding involved in DNA replication; P:DNA replication initiation; P:mitotic DNA replication; F:single-stranded DNA binding; F:ATP binding; F:ATP hydrolysis activity; F:single-stranded 3 -5 DNA helicase activity; C:THO complex; C:MCM complex | Acting on acid anhydrides; DNA helicase | P:DNA duplex unwinding; F:DNA binding; F:ATP binding | view details | ||
| JG649261.1 | Tinospora cordifolia | putative exosome complex component rrp40 | 644 | P:GO:0000467; P:GO:0034427; P:GO:0034475; P:GO:0043928; P:GO:0071028; P:GO:0071034; P:GO:0071035; P:GO:0071038; P:GO:0071051; F:GO:0003723; F:GO:0004527; C:GO:0000176; C:GO:0000177 | P:exonucleolytic trimming to generate mature 3 -end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); P:nuclear-transcribed mRNA catabolic process, exonucleolytic, 3 -5; P:U4 snRNA 3 -end processing; P:exonucleolytic catabolism of deadenylated mRNA; P:nuclear mRNA surveillance; P:CUT catabolic process; P:nuclear polyadenylation-dependent rRNA catabolic process; P:nuclear polyadenylation-dependent tRNA catabolic process; P:polyadenylation-dependent snoRNA 3-end processing; F:RNA binding; F:exonuclease activity; C:nuclear exosome (RNase complex); C:cytoplasmic exosome (RNase complex) | Acting on ester bonds | F:RNA binding; C:exosome (RNase complex) | view details | ||
| JG650623.1 | Tinospora cordifolia | bifunctional purine biosynthesis protein PurH | 725 | P:GO:0006189; F:GO:0003937; F:GO:0004643; C:GO:0005829 | P:de novo IMP biosynthetic process; F:IMP cyclohydrolase activity; F:phosphoribosylaminoimidazolecarboxamide formyltransferase activity; C:cytosol | IMP cyclohydrolase; Phosphoribosylaminoimidazolecarboxamide formyltransferase | P:purine nucleotide biosynthetic process; F:IMP cyclohydrolase activity; F:phosphoribosylaminoimidazolecarboxamide formyltransferase activity | view details | ||
| JG650022.1 | Tinospora cordifolia | putative CCA tRNA nucleotidyltransferase 2 isoform X1 | 750 | P:GO:0001680; F:GO:0003723; F:GO:0016437; F:GO:0052927; F:GO:0052928; F:GO:0052929 | P:tRNA 3-terminal CCA addition; F:RNA binding; F:tRNA cytidylyltransferase activity; F:CTP:tRNA cytidylyltransferase activity; F:CTP:3-cytidine-tRNA cytidylyltransferase activity; F:ATP:3-cytidine-cytidine-tRNA adenylyltransferase activity | CCA tRNA nucleotidyltransferase; Polynucleotide adenylyltransferase | P:RNA processing; F:RNA binding; F:nucleotidyltransferase activity | view details | ||
| JG650113.1 | Tinospora cordifolia | putative CCA tRNA nucleotidyltransferase 2 | 755 | P:GO:0001680; F:GO:0003723; F:GO:0016437; F:GO:0052927; F:GO:0052928; F:GO:0052929 | P:tRNA 3-terminal CCA addition; F:RNA binding; F:tRNA cytidylyltransferase activity; F:CTP:tRNA cytidylyltransferase activity; F:CTP:3 -cytidine-tRNA cytidylyltransferase activity; F:ATP:3 -cytidine-cytidine-tRNA adenylyltransferase activity | CCA tRNA nucleotidyltransferase; Polynucleotide adenylyltransferase | P:RNA processing; F:RNA binding; F:nucleotidyltransferase activity | view details | ||
| JG646478.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 698 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645977.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 591 | F:protein binding | view details | |||||
| JG649578.1 | Tinospora cordifolia | Bowman-Birk type proteinase inhibitor-like isoform X2 | 807 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:serine-type endopeptidase inhibitor activity; C:extracellular region | view details | |||
| JG648419.1 | Tinospora cordifolia | thaumatin-like protein | 835 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648251.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 562 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | F:protein binding | view details | |||
| JG650453.1 | Tinospora cordifolia | probable glutathione S-transferase | 804 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG645747.1 | Tinospora cordifolia | 60S ribosomal protein L23 | 590 | P:GO:0006412; F:GO:0003735; F:GO:0070180; C:GO:0022625 | P:translation; F:structural constituent of ribosome; F:large ribosomal subunit rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647861.1 | Tinospora cordifolia | actin-depolymerizing factor | 798 | P:GO:0030042; F:GO:0051015; C:GO:0005737; C:GO:0015629 | P:actin filament depolymerization; F:actin filament binding; C:cytoplasm; C:actin cytoskeleton | P:actin filament depolymerization; F:actin binding; C:actin cytoskeleton | view details | |||
| JG648667.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 807 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648945.1 | Tinospora cordifolia | major strawberry allergen Fra a 1-2-like | 644 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648930.1 | Tinospora cordifolia | ripening-related protein grip22 | 794 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646689.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 800 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG651009.1 | Tinospora cordifolia | uncharacterized protein LOC122072982 | 804 | no GO terms | view details | |||||
| JG647126.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 812 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648818.1 | Tinospora cordifolia | protein CREG1-like | 772 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646788.1 | Tinospora cordifolia | Putative DNA repair protein RAD23-3 | 552 | P:GO:0006289; P:GO:0043161; F:GO:0003684; F:GO:0031593; F:GO:0043130; F:GO:0070628; C:GO:0005654; C:GO:0005829 | P:nucleotide-excision repair; P:proteasome-mediated ubiquitin-dependent protein catabolic process; F:damaged DNA binding; F:polyubiquitin modification-dependent protein binding; F:ubiquitin binding; F:proteasome binding; C:nucleoplasm; C:cytosol | P:nucleotide-excision repair; P:proteasome-mediated ubiquitin-dependent protein catabolic process; F:damaged DNA binding; F:protein binding | view details | |||
| JG647212.1 | Tinospora cordifolia | pheophorbide a oxygenase, chloroplastic | 805 | F:GO:0010277; F:GO:0032441; F:GO:0046872; F:GO:0051537; C:GO:0009507; C:GO:0031967 | F:chlorophyllide a oxygenase [overall] activity; F:pheophorbide a oxygenase activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; C:chloroplast; C:organelle envelope | Acting on single donors with incorporation of molecular oxygen (oxygenases). The oxygen incorporated need not be derived from O(2); Chlorophyllide a oxygenase; Acting on iron-sulfur proteins as donors; Pheophorbide a oxygenase | F:2 iron, 2 sulfur cluster binding | view details | ||
| JG647607.1 | Tinospora cordifolia | embryo-specific protein ATS3A-like | 812 | C:GO:0005737 | C:cytoplasm | F:protein binding | view details | |||
| JG648127.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 818 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG649467.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 816 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650114.1 | Tinospora cordifolia | ADP-ribosylation factor 2 isoform X1 | 779 | F:GO:0003924; F:GO:0005525 | F:GTPase activity; F:GTP binding | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG646494.1 | Tinospora cordifolia | B2 protein | 834 | P:GO:0034976 | P:response to endoplasmic reticulum stress | P:response to endoplasmic reticulum stress | view details | |||
| JG650371.1 | Tinospora cordifolia | thaumatin-like protein | 803 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG647811.1 | Tinospora cordifolia | autophagy-related protein 8f | 691 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016020 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:membrane | no GO terms | view details | |||
| JG650666.1 | Tinospora cordifolia | N-(5-amino-5-carboxypentanoyl)-L-cysteinyl-D-valine synthase | 819 | F:GO:0003676 | F:nucleic acid binding | F:nucleic acid binding | view details | |||
| JG650562.1 | Tinospora cordifolia | AP-1 complex subunit sigma-2 | 867 | P:GO:0006886; P:GO:0016192; F:GO:0035615; C:GO:0030121 | P:intracellular protein transport; P:vesicle-mediated transport; F:clathrin adaptor activity; C:AP-1 adaptor complex | P:protein transport; P:vesicle-mediated transport; F:clathrin adaptor activity; C:AP-1 adaptor complex | view details | |||
| JG649227.1 | Tinospora cordifolia | thaumatin-like protein | 797 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG648203.1 | Tinospora cordifolia | dirigent protein 22-like | 698 | C:GO:0016021; C:GO:0048046 | C:integral component of membrane; C:apoplast | no GO terms | view details | |||
| JG648217.1 | Tinospora cordifolia | uricase-2 isozyme 2 | 514 | P:GO:0006145; P:GO:0019628; F:GO:0004846; C:GO:0005777 | P:purine nucleobase catabolic process; P:urate catabolic process; F:urate oxidase activity; C:peroxisome | Factor independent urate hydroxylase | no GO terms | view details | ||
| JG650144.1 | Tinospora cordifolia | putative disease resistance protein RGA4 | 589 | P:GO:0006952; P:GO:0006955; P:GO:0098542; F:GO:0043531 | P:defense response; P:immune response; P:defense response to other organism; F:ADP binding | no GO terms | view details | |||
| JG649535.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 664 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648628.1 | Tinospora cordifolia | hypothetical protein BVC80_9089g103 | 690 | no GO terms | view details | |||||
| JG649336.1 | Tinospora cordifolia | glutathione S-transferase F9-like | 736 | P:GO:0006749; P:GO:0009407; F:GO:0004364; F:GO:0043295; C:GO:0005829 | P:glutathione metabolic process; P:toxin catabolic process; F:glutathione transferase activity; F:glutathione binding; C:cytosol | Glutathione transferase | no GO terms | view details | ||
| JG649520.1 | Tinospora cordifolia | thaumatin-like protein | 840 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG646053.1 | Tinospora cordifolia | Hypoxia induced protein, domain containing protein | 689 | P:GO:0033617; C:GO:0005746; C:GO:0031305 | P:mitochondrial cytochrome c oxidase assembly; C:mitochondrial respirasome; C:integral component of mitochondrial inner membrane | no GO terms | view details | |||
| JG647680.1 | Tinospora cordifolia | 40S ribosomal protein S23 | 569 | P:GO:0006412; F:GO:0003735; C:GO:0016021; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:integral component of membrane; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG647242.1 | Tinospora cordifolia | Bet v I domain | 803 | P:GO:0006952 | P:defense response | P:defense response | view details | |||
| JG646210.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 646 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646470.1 | Tinospora cordifolia | enolase | 805 | P:GO:0006096; F:GO:0000287; F:GO:0004634; C:GO:0000015 | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | Phosphopyruvate hydratase | P:glycolytic process; F:magnesium ion binding; F:phosphopyruvate hydratase activity; C:phosphopyruvate hydratase complex | view details | ||
| JG647329.1 | Tinospora cordifolia | constitutive photomorphogenesis protein 10 | 577 | P:GO:0000209; P:GO:0010099; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:regulation of photomorphogenesis; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG648923.1 | Tinospora cordifolia | gibberellin-regulated protein 1-like | 602 | P:GO:0009737; P:GO:0009740; P:GO:0009741; C:GO:0005576; C:GO:0016021 | P:response to abscisic acid; P:gibberellic acid mediated signaling pathway; P:response to brassinosteroid; C:extracellular region; C:integral component of membrane | no GO terms | view details | |||
| JG648143.1 | Tinospora cordifolia | glutamate dehydrogenase 2 | 631 | P:GO:0006538; F:GO:0000166; F:GO:0004352; C:GO:0005739 | P:glutamate catabolic process; F:nucleotide binding; F:glutamate dehydrogenase (NAD+) activity; C:mitochondrion | Glutamate dehydrogenase (NAD(P)(+)); Glutamate dehydrogenase | P:cellular amino acid metabolic process; F:oxidoreductase activity | view details | ||
| JG650845.1 | Tinospora cordifolia | 60S ribosomal protein L12 | 646 | P:GO:0006412; F:GO:0003735; F:GO:0070180; C:GO:0022625 | P:translation; F:structural constituent of ribosome; F:large ribosomal subunit rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650693.1 | Tinospora cordifolia | chaperone protein ClpD, chloroplastic-like | 567 | P:GO:0065003; F:GO:0005524; F:GO:0016887; C:GO:0009570 | P:protein-containing complex assembly; F:ATP binding; F:ATP hydrolysis activity; C:chloroplast stroma | Nucleoside-triphosphate phosphatase | no GO terms | view details | ||
| JG646595.1 | Tinospora cordifolia | alpha-amylase/subtilisin inhibitor-like | 730 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647058.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 807 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646498.1 | Tinospora cordifolia | aromatic aminotransferase ISS1 | 696 | P:GO:0006520; P:GO:1901566; F:GO:0004069; F:GO:0030170 | P:cellular amino acid metabolic process; P:organonitrogen compound biosynthetic process; F:L-aspartate:2-oxoglutarate aminotransferase activity; F:pyridoxal phosphate binding | Aspartate transaminase | P:biosynthetic process; F:catalytic activity; F:pyridoxal phosphate binding | view details | ||
| JG648662.1 | Tinospora cordifolia | protein TIFY 3B | 847 | P:GO:0048583 | P:regulation of response to stimulus | no GO terms | view details | |||
| JG647089.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 804 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648085.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2-17 kDa | 780 | P:GO:0000209; P:GO:0006511; F:GO:0004839; F:GO:0005524; F:GO:0016746; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ubiquitin activating enzyme activity; F:ATP binding; F:acyltransferase activity; F:ubiquitin conjugating enzyme activity; C:nucleus | E1 ubiquitin-activating enzyme; E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG650359.1 | Tinospora cordifolia | ---NA--- | 774 | P:plant-type cell wall organization; F:structural constituent of cell wall | view details | |||||
| JG649415.1 | Tinospora cordifolia | pathogenesis-related protein STH-21-like | 753 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648468.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 801 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650638.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 800 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648457.1 | Tinospora cordifolia | ripening-related protein grip22 | 767 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648653.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 805 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646168.1 | Tinospora cordifolia | ripening-related protein grip22 | 812 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646215.1 | Tinospora cordifolia | hypothetical protein HHK36_026134 | 729 | F:GO:0046872 | F:metal ion binding | no GO terms | view details | |||
| JG646027.1 | Tinospora cordifolia | pollen-specific protein C13-like | 720 | C:GO:0005615 | C:extracellular space | no GO terms | view details | |||
| JG647085.1 | Tinospora cordifolia | 60S ribosomal protein L11-1 | 788 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648692.1 | Tinospora cordifolia | light-harvesting complex-like protein OHP2, chloroplastic | 814 | P:GO:0006850; P:GO:0015979; P:GO:0016310; F:GO:0016301; F:GO:0046872; F:GO:0050833; C:GO:0009507; C:GO:0009579; C:GO:0031305 | P:mitochondrial pyruvate transmembrane transport; P:photosynthesis; P:phosphorylation; F:kinase activity; F:metal ion binding; F:pyruvate transmembrane transporter activity; C:chloroplast; C:thylakoid; C:integral component of mitochondrial inner membrane | Transferring phosphorus-containing groups; Translocases | no GO terms | view details | ||
| JG646060.1 | Tinospora cordifolia | probable serine/threonine-protein kinase PBL3 | 786 | P:GO:0006468; F:GO:0004674; F:GO:0005524 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG648837.1 | Tinospora cordifolia | plastid-lipid-associated protein, chloroplastic-like | 796 | no GO terms | view details | |||||
| JG648919.1 | Tinospora cordifolia | metallothiol transferase FosB | 817 | no GO terms | view details | |||||
| JG649477.1 | Tinospora cordifolia | protein yippee-like | 778 | C:GO:0000151 | C:ubiquitin ligase complex | no GO terms | view details | |||
| JG647460.1 | Tinospora cordifolia | octicosapeptide/Phox/Bem1p (PB1) domain-containing protein | 797 | P:GO:0016310; F:GO:0016301 | P:phosphorylation; F:kinase activity | F:protein binding | view details | |||
| JG650115.1 | Tinospora cordifolia | membrane steroid-binding protein 2-like | 754 | C:GO:0005783; C:GO:0016021 | C:endoplasmic reticulum; C:integral component of membrane | no GO terms | view details | |||
| JG646001.1 | Tinospora cordifolia | Burp domain protein rd22 | 818 | no GO terms | view details | |||||
| JG650450.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 677 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645761.1 | Tinospora cordifolia | thaumatin-like protein | 771 | no GO terms | view details | |||||
| JG648772.1 | Tinospora cordifolia | D-lactate dehydrogenase [cytochrome], mitochondrial isoform X1 | 822 | P:GO:1903457; F:GO:0004458; F:GO:0008720; F:GO:0071949; C:GO:0005739 | P:lactate catabolic process; F:D-lactate dehydrogenase (cytochrome) activity; F:D-lactate dehydrogenase activity; F:FAD binding; C:mitochondrion | D-lactate dehydrogenase (cytochrome); D-lactate dehydrogenase | F:flavin adenine dinucleotide binding; F:FAD binding | view details | ||
| NC_042153.1 | Tinospora cordifolia | 150945 | F:ATP binding; F:ATP hydrolysis activity | view details | ||||||
| JG645929.1 | Tinospora cordifolia | polyphenol oxidase I, chloroplastic-like | 462 | P:GO:0046148; F:GO:0004097; F:GO:0004503; F:GO:0046872; C:GO:0016021 | P:pigment biosynthetic process; F:catechol oxidase activity; F:tyrosinase activity; F:metal ion binding; C:integral component of membrane | Tyrosinase; Catechol oxidase | F:catechol oxidase activity | view details | ||
| JG648434.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 750 | F:protein binding | view details | |||||
| JG647935.1 | Tinospora cordifolia | calmodulin-like protein 8 | 637 | F:GO:0005509; F:GO:0032440 | F:calcium ion binding; F:2-alkenal reductase [NAD(P)+] activity | 2-alkenal reductase (NAD(P)(+)) | F:calcium ion binding | view details | ||
| JG650234.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 673 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646322.1 | Tinospora cordifolia | serine/arginine repetitive matrix protein 1-like isoform X1 | 867 | P:GO:0006397 | P:mRNA processing | no GO terms | view details | |||
| JG647646.1 | Tinospora cordifolia | ---NA--- | 748 | no GO terms | view details | |||||
| JG650207.1 | Tinospora cordifolia | RNA recognition motif domain | 535 | F:GO:0003723; C:GO:0016021 | F:RNA binding; C:integral component of membrane | F:nucleic acid binding; F:RNA binding | view details | |||
| JG647716.1 | Tinospora cordifolia | DUF1685 domain-containing protein | 599 | no GO terms | view details | |||||
| JG646111.1 | Tinospora cordifolia | glycosyltransferase BC10-like | 757 | F:GO:0016757; C:GO:0016021 | F:glycosyltransferase activity; C:integral component of membrane | Glycosyltransferases | F:glycosyltransferase activity; C:membrane | view details | ||
| JG650544.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 793 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650654.1 | Tinospora cordifolia | lipid transfer-like protein vas | 739 | C:GO:0016020 | C:membrane | P:lipid transport; F:lipid binding | view details | |||
| JG645827.1 | Tinospora cordifolia | protein C2-DOMAIN ABA-RELATED 1-like | 660 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG647470.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 679 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650712.1 | Tinospora cordifolia | Glyoxalase-like domain | 768 | no GO terms | view details | |||||
| JG647169.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 796 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650317.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 581 | F:protein binding | view details | |||||
| JG646415.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 721 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647435.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 799 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648207.1 | Tinospora cordifolia | putative E3 ubiquitin-protein ligase XBAT35 | 794 | F:GO:0016740; F:GO:0140096 | F:transferase activity; F:catalytic activity, acting on a protein | Transferases | F:ubiquitin-protein transferase activity | view details | ||
| JG649367.1 | Tinospora cordifolia | ETHYLENE INSENSITIVE 3-like 1 protein | 776 | P:GO:0050794; F:GO:0003677; F:GO:0003700 | P:regulation of cellular process; F:DNA binding; F:DNA-binding transcription factor activity | C:nucleus | view details | |||
| JG646785.1 | Tinospora cordifolia | protein SMG7-like | 781 | P:GO:0000184; F:GO:0042162; F:GO:0070034; C:GO:0005697 | P:nuclear-transcribed mRNA catabolic process, nonsense-mediated decay; F:telomeric DNA binding; F:telomerase RNA binding; C:telomerase holoenzyme complex | no GO terms | view details | |||
| JG649162.1 | Tinospora cordifolia | Hypothetical predicted protein | 634 | C:GO:0005737; C:GO:0005794 | C:cytoplasm; C:Golgi apparatus | no GO terms | view details | |||
| JG647835.1 | Tinospora cordifolia | probable glutathione S-transferase | 793 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG648091.1 | Tinospora cordifolia | kunitz trypsin inhibitor 5-like | 783 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649085.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 602 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG649377.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A-2 | 816 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG647173.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 585 | F:protein binding | view details | |||||
| JG650024.1 | Tinospora cordifolia | membrane steroid-binding protein 2-like | 757 | C:GO:0005783; C:GO:0016021 | C:endoplasmic reticulum; C:integral component of membrane | no GO terms | view details | |||
| JG647573.1 | Tinospora cordifolia | prohibitin-1, mitochondrial | 859 | P:GO:0007005; C:GO:0005743; C:GO:0005886 | P:mitochondrion organization; C:mitochondrial inner membrane; C:plasma membrane | C:membrane | view details | |||
| JG645997.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 681 | F:protein binding | view details | |||||
| JG648145.1 | Tinospora cordifolia | probable calcium-binding protein CML36 | 803 | F:GO:0005509 | F:calcium ion binding | F:calcium ion binding | view details | |||
| JG647833.1 | Tinospora cordifolia | ADP-ribosylation factor-like | 800 | P:GO:0006886; P:GO:0016192; F:GO:0003924; F:GO:0005525; C:GO:0009536 | P:intracellular protein transport; P:vesicle-mediated transport; F:GTPase activity; F:GTP binding; C:plastid | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG648166.1 | Tinospora cordifolia | cell division topological specificity factor homolog, chloroplastic | 809 | P:GO:0010020; F:GO:0005515; C:GO:0009507 | P:chloroplast fission; F:protein binding; C:chloroplast | P:regulation of division septum assembly; P:cell division | view details | |||
| JG650714.1 | Tinospora cordifolia | protein CREG1 | 754 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650462.1 | Tinospora cordifolia | protein CURVATURE THYLAKOID 1A, chloroplastic-like | 706 | C:GO:0009535; C:GO:0016021 | C:chloroplast thylakoid membrane; C:integral component of membrane | C:thylakoid | view details | |||
| JG650648.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 596 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG646907.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 579 | F:protein binding | view details | |||||
| JG648601.1 | Tinospora cordifolia | probable glutathione S-transferase | 823 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG648481.1 | Tinospora cordifolia | thaumatin-like protein | 783 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648828.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 801 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649046.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 798 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646148.1 | Tinospora cordifolia | tRNA (cytosine(38)-C(5))-methyltransferase 2 | 820 | P:GO:0032259; F:GO:0008168 | P:methylation; F:methyltransferase activity | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG649837.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 782 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648545.1 | Tinospora cordifolia | peptidyl-prolyl cis-trans isomerase isoform X2 | 734 | P:GO:0000413; P:GO:0006457; F:GO:0003755; F:GO:0016018; C:GO:0005737 | P:protein peptidyl-prolyl isomerization; P:protein folding; F:peptidyl-prolyl cis-trans isomerase activity; F:cyclosporin A binding; C:cytoplasm | Peptidylprolyl isomerase | P:protein peptidyl-prolyl isomerization; F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG650281.1 | Tinospora cordifolia | alpha-amylase/subtilisin inhibitor-like | 738 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650330.1 | Tinospora cordifolia | iron-sulfur assembly protein IscA-like 1, mitochondrial | 724 | P:GO:0016226; P:GO:0097428; F:GO:0051537; C:GO:0005739 | P:iron-sulfur cluster assembly; P:protein maturation by iron-sulfur cluster transfer; F:2 iron, 2 sulfur cluster binding; C:mitochondrion | P:iron-sulfur cluster assembly; F:iron-sulfur cluster binding | view details | |||
| JG647540.1 | Tinospora cordifolia | neutral/alkaline invertase 3, chloroplastic-like | 692 | P:GO:0005982; P:GO:0005987; P:GO:0048825; F:GO:0004575; F:GO:0033926; C:GO:0009507 | P:starch metabolic process; P:sucrose catabolic process; P:cotyledon development; F:sucrose alpha-glucosidase activity; F:glycopeptide alpha-N-acetylgalactosaminidase activity; C:chloroplast | Endo-alpha-N-acetylgalactosaminidase; Alpha-glucosidase; Beta-fructofuranosidase; Sucrose alpha-glucosidase | P:carbohydrate metabolic process; F:glycopeptide alpha-N-acetylgalactosaminidase activity | view details | ||
| JG649301.1 | Tinospora cordifolia | thaumatin-like protein | 749 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650824.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 795 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648705.1 | Tinospora cordifolia | putative germin-like protein 2-1 | 837 | F:GO:0030145; C:GO:0048046 | F:manganese ion binding; C:apoplast | F:manganese ion binding | view details | |||
| JG646635.1 | Tinospora cordifolia | D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like | 766 | P:GO:0006564; F:GO:0004617; F:GO:0051287; C:GO:0009570 | P:L-serine biosynthetic process; F:phosphoglycerate dehydrogenase activity; F:NAD binding; C:chloroplast stroma | Phosphoglycerate dehydrogenase | no GO terms | view details | ||
| JG646800.1 | Tinospora cordifolia | probable calcium-binding protein CML18 | 704 | F:GO:0005509 | F:calcium ion binding | F:calcium ion binding | view details | |||
| JG648894.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 585 | F:protein binding | view details | |||||
| JG647695.1 | Tinospora cordifolia | 60S ribosomal protein L34 | 655 | P:GO:0006412; P:GO:0042254; F:GO:0003735; C:GO:0022625 | P:translation; P:ribosome biogenesis; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650375.1 | Tinospora cordifolia | triosephosphate isomerase, cytosolic | 547 | P:GO:0006094; P:GO:0006096; P:GO:0010043; P:GO:0019563; P:GO:0046166; F:GO:0004807; C:GO:0005739; C:GO:0005829 | P:gluconeogenesis; P:glycolytic process; P:response to zinc ion; P:glycerol catabolic process; P:glyceraldehyde-3-phosphate biosynthetic process; F:triose-phosphate isomerase activity; C:mitochondrion; C:cytosol | Triose-phosphate isomerase | F:triose-phosphate isomerase activity | view details | ||
| JG648735.1 | Tinospora cordifolia | guanosine deaminase | 745 | P:GO:0006152; F:GO:0008270; F:GO:0047974 | P:purine nucleoside catabolic process; F:zinc ion binding; F:guanosine deaminase activity | Guanosine deaminase | F:catalytic activity | view details | ||
| JG646857.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 577 | P:GO:0005975; F:GO:0016765; F:GO:0042973 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647984.1 | Tinospora cordifolia | subtilisin-like protease SBT3.17 | 594 | P:GO:0006508; P:GO:0016310; F:GO:0008233; F:GO:0016301 | P:proteolysis; P:phosphorylation; F:peptidase activity; F:kinase activity | Acting on peptide bonds (peptidases); Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG649655.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 600 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG646231.1 | Tinospora cordifolia | Sap domain | 607 | P:GO:0032436; C:GO:0080008 | P:positive regulation of proteasomal ubiquitin-dependent protein catabolic process; C:Cul4-RING E3 ubiquitin ligase complex | P:regulation of proteasomal ubiquitin-dependent protein catabolic process | view details | |||
| JG647002.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 603 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG649393.1 | Tinospora cordifolia | N-terminal acetyltransferase B complex catalytic subunit NAA20 | 675 | P:GO:0017196; F:GO:0004596; C:GO:0031416 | P:N-terminal peptidyl-methionine acetylation; F:peptide alpha-N-acetyltransferase activity; C:NatB complex | Acyltransferases | F:acetyltransferase activity | view details | ||
| JG646175.1 | Tinospora cordifolia | berberine bridge enzyme | 650 | F:GO:0016491; F:GO:0071949; C:GO:0005634 | F:oxidoreductase activity; F:FAD binding; C:nucleus | Oxidoreductases | F:flavin adenine dinucleotide binding | view details | ||
| JG647693.1 | Tinospora cordifolia | kunitz trypsin inhibitor 5-like | 797 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648626.1 | Tinospora cordifolia | alpha-amylase/subtilisin inhibitor-like | 740 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645814.1 | Tinospora cordifolia | ripening-related protein grip22 | 770 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649693.1 | Tinospora cordifolia | non-specific lipid-transfer protein 2-like | 563 | P:GO:0006869 | P:lipid transport | P:lipid transport | view details | |||
| JG648866.1 | Tinospora cordifolia | major strawberry allergen Fra a 1-2-like | 707 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645984.1 | Tinospora cordifolia | protein C2-DOMAIN ABA-RELATED 4-like | 717 | P:GO:0009738; C:GO:0005634; C:GO:0005886 | P:abscisic acid-activated signaling pathway; C:nucleus; C:plasma membrane | no GO terms | view details | |||
| JG649352.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 600 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG646594.1 | Tinospora cordifolia | 40S ribosomal protein S11 | 755 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649340.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 741 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649995.1 | Tinospora cordifolia | 60S ribosomal protein L23A | 625 | P:GO:0000027; P:GO:0006412; F:GO:0003729; F:GO:0003735; F:GO:0019843; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:mRNA binding; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647526.1 | Tinospora cordifolia | non-specific lipid-transfer protein 2-like | 563 | P:GO:0006869 | P:lipid transport | P:lipid transport | view details | |||
| JG645791.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 600 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG646512.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 707 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649369.1 | Tinospora cordifolia | thioredoxin-like protein Clot | 699 | P:GO:0098869; F:GO:0004601; F:GO:0047134; C:GO:0005829 | P:cellular oxidant detoxification; F:peroxidase activity; F:protein-disulfide reductase (NAD(P)) activity; C:cytosol | Acting on a peroxide as acceptor; Protein-disulfide reductase | F:protein-disulfide reductase (NAD(P)) activity | view details | ||
| JG647447.1 | Tinospora cordifolia | ETHYLENE INSENSITIVE 3-like 3 protein isoform X1 | 840 | P:GO:0006355; P:GO:0009873; F:GO:0003677; F:GO:0003700; C:GO:0005634 | P:regulation of DNA-templated transcription; P:ethylene-activated signaling pathway; F:DNA binding; F:DNA-binding transcription factor activity; C:nucleus | C:nucleus | view details | |||
| JG649381.1 | Tinospora cordifolia | protein CREG1-like | 722 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648138.1 | Tinospora cordifolia | calcium load-activated calcium channel-like | 826 | P:GO:0032469; P:GO:0070588; F:GO:0005262; C:GO:0030176 | P:endoplasmic reticulum calcium ion homeostasis; P:calcium ion transmembrane transport; F:calcium channel activity; C:integral component of endoplasmic reticulum membrane | Translocases | P:endoplasmic reticulum calcium ion homeostasis; F:calcium channel activity; C:membrane; C:integral component of endoplasmic reticulum membrane | view details | ||
| JG645935.1 | Tinospora cordifolia | At-rich interactive domain protein | 817 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG649466.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 791 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646232.1 | Tinospora cordifolia | pathogenesis-related protein 1-like | 638 | C:GO:0005615 | C:extracellular space | no GO terms | view details | |||
| JG650958.1 | Tinospora cordifolia | 60S ribosomal protein L24 | 692 | P:GO:0002181; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | no GO terms | view details | |||
| JG650286.1 | Tinospora cordifolia | protein CREG1-like | 759 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646555.1 | Tinospora cordifolia | ORM1-like protein 1 | 806 | P:GO:0090156; P:GO:1900060; C:GO:0016021; C:GO:0035339 | P:cellular sphingolipid homeostasis; P:negative regulation of ceramide biosynthetic process; C:integral component of membrane; C:SPOTS complex | C:endoplasmic reticulum membrane; C:integral component of membrane | view details | |||
| JG649533.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 799 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647785.1 | Tinospora cordifolia | universal stress protein PHOS32 | 771 | no GO terms | view details | |||||
| JG646073.1 | Tinospora cordifolia | 40S ribosomal protein S24-1 | 721 | P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0005829; C:GO:0005840 | P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosol; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646346.1 | Tinospora cordifolia | dihydroneopterin aldolase 2 isoform X1 | 742 | P:GO:0046654; P:GO:0046656; F:GO:0004150; C:GO:0005737 | P:tetrahydrofolate biosynthetic process; P:folic acid biosynthetic process; F:dihydroneopterin aldolase activity; C:cytoplasm | Dihydroneopterin aldolase | P:folic acid-containing compound metabolic process; F:dihydroneopterin aldolase activity | view details | ||
| JG646965.1 | Tinospora cordifolia | probable mediator of RNA polymerase II transcription subunit 37c | 504 | F:GO:0005524; F:GO:0016887 | F:ATP binding; F:ATP hydrolysis activity | Nucleoside-triphosphate phosphatase | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG649790.1 | Tinospora cordifolia | glutathione S-transferase F9-like | 806 | P:GO:0006749; P:GO:0009407; F:GO:0004364; F:GO:0043295; C:GO:0005829 | P:glutathione metabolic process; P:toxin catabolic process; F:glutathione transferase activity; F:glutathione binding; C:cytosol | Glutathione transferase | no GO terms | view details | ||
| JG647387.1 | Tinospora cordifolia | thaumatin-like protein | 835 | no GO terms | view details | |||||
| JG648905.1 | Tinospora cordifolia | macrophage migration inhibitory factor homolog isoform X1 | 633 | F:GO:0050178; C:GO:0005615; C:GO:0005634; C:GO:0005737 | F:phenylpyruvate tautomerase activity; C:extracellular space; C:nucleus; C:cytoplasm | Phenylpyruvate tautomerase | no GO terms | view details | ||
| JG646525.1 | Tinospora cordifolia | thaumatin-like protein | 826 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG647678.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein LOC104597634 | 782 | no GO terms | view details | |||||
| JG647188.1 | Tinospora cordifolia | probable glutathione S-transferase | 791 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG650059.1 | Tinospora cordifolia | thaumatin-like protein | 793 | no GO terms | view details | |||||
| JG647981.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 variant 1D | 672 | P:GO:0006301; P:GO:0070534; C:GO:0005634 | P:postreplication repair; P:protein K63-linked ubiquitination; C:nucleus | no GO terms | view details | |||
| JG649646.1 | Tinospora cordifolia | proteasome subunit beta type-2-A | 820 | P:GO:0010498; F:GO:0004298; C:GO:0005634; C:GO:0005737; C:GO:0019774 | P:proteasomal protein catabolic process; F:threonine-type endopeptidase activity; C:nucleus; C:cytoplasm; C:proteasome core complex, beta-subunit complex | Acting on peptide bonds (peptidases) | P:proteasomal protein catabolic process; P:proteolysis involved in protein catabolic process; C:proteasome core complex | view details | ||
| JG649570.1 | Tinospora cordifolia | serine/threonine-protein kinase PBS1-like | 719 | P:GO:0006468; F:GO:0004674; F:GO:0005524 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG648268.1 | Tinospora cordifolia | peptidyl-prolyl cis-trans isomerase | 671 | P:GO:0000413; P:GO:0006457; F:GO:0003755; F:GO:0016018; C:GO:0005737 | P:protein peptidyl-prolyl isomerization; P:protein folding; F:peptidyl-prolyl cis-trans isomerase activity; F:cyclosporin A binding; C:cytoplasm | Peptidylprolyl isomerase | P:protein peptidyl-prolyl isomerization; F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG648003.1 | Tinospora cordifolia | protein disulfide isomerase pTAC5, chloroplastic | 706 | P:GO:0006807; P:GO:0009628; P:GO:0009658; P:GO:0043170; P:GO:0044238; F:GO:0003756; C:GO:0009507 | P:nitrogen compound metabolic process; P:response to abiotic stimulus; P:chloroplast organization; P:macromolecule metabolic process; P:primary metabolic process; F:protein disulfide isomerase activity; C:chloroplast | Protein disulfide-isomerase | no GO terms | view details | ||
| JG648624.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 596 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG645570.1 | Tinospora cordifolia | protein PLANT CADMIUM RESISTANCE 8 | 698 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650622.1 | Tinospora cordifolia | Splicing factor 3B subunit 4 | 793 | F:GO:0003676 | F:nucleic acid binding | F:nucleic acid binding; F:RNA binding | view details | |||
| JG648356.1 | Tinospora cordifolia | glutathione S-transferase U17-like | 635 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG647272.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 720 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646567.1 | Tinospora cordifolia | cytochrome P450 | 634 | P:GO:0009699; P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:phenylpropanoid biosynthetic process; P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646418.1 | Tinospora cordifolia | probable glutathione S-transferase | 798 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG645916.1 | Tinospora cordifolia | non-specific lipid-transfer protein 2-like | 566 | P:GO:0006869 | P:lipid transport | P:lipid transport | view details | |||
| JG650091.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 691 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG647062.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 639 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645592.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 670 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649126.1 | Tinospora cordifolia | 60S ribosomal protein L15 | 709 | P:GO:0002181; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650631.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 643 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG650971.1 | Tinospora cordifolia | thaumatin-like protein | 829 | no GO terms | view details | |||||
| JG649762.1 | Tinospora cordifolia | MLO-like protein 6 | 663 | P:GO:0006952; P:GO:0009607; F:GO:0005516; C:GO:0016021 | P:defense response; P:response to biotic stimulus; F:calmodulin binding; C:integral component of membrane | P:defense response; C:integral component of membrane | view details | |||
| JG650167.1 | Tinospora cordifolia | translin isoform X1 | 755 | P:GO:0016070; F:GO:0003697; F:GO:0003723; F:GO:0043565; C:GO:0005634; C:GO:0005737 | P:RNA metabolic process; F:single-stranded DNA binding; F:RNA binding; F:sequence-specific DNA binding; C:nucleus; C:cytoplasm | P:RNA metabolic process; F:single-stranded DNA binding; F:RNA binding; F:sequence-specific DNA binding | view details | |||
| JG647880.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 800 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649607.1 | Tinospora cordifolia | thaumatin-like protein | 812 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648159.1 | Tinospora cordifolia | ADP-ribosylation factor-like protein 8c | 628 | P:GO:0015031; F:GO:0003924; F:GO:0005525; C:GO:0005774 | P:protein transport; F:GTPase activity; F:GTP binding; C:vacuolar membrane | Nucleoside-triphosphate phosphatase | P:protein transport; F:GTPase activity; F:GTP binding | view details | ||
| JG649821.1 | Tinospora cordifolia | putative L-ascorbate peroxidase 6 isoform X1 | 759 | P:GO:0006979; P:GO:0070887; F:GO:0004601 | P:response to oxidative stress; P:cellular response to chemical stimulus; F:peroxidase activity | Acting on a peroxide as acceptor | P:response to oxidative stress; P:cellular response to oxidative stress; F:peroxidase activity; F:heme binding | view details | ||
| JG646890.1 | Tinospora cordifolia | thaumatin-like protein | 736 | no GO terms | view details | |||||
| JG649490.1 | Tinospora cordifolia | 60S ribosomal protein L9 | 797 | P:GO:0002181; F:GO:0003735; F:GO:0019843; C:GO:0022625 | P:cytoplasmic translation; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:ribosome | view details | |||
| JG648798.1 | Tinospora cordifolia | hypothetical protein HHK36_020759 | 519 | P:GO:0045892; F:GO:0003714; C:GO:0005634 | P:negative regulation of DNA-templated transcription; F:transcription corepressor activity; C:nucleus | no GO terms | view details | |||
| JG650440.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 782 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646698.1 | Tinospora cordifolia | Mannose-binding lectin | 656 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG645650.1 | Tinospora cordifolia | polyphenol oxidase I, chloroplastic-like | 615 | P:GO:0046148; F:GO:0004097; F:GO:0004503; F:GO:0046872; C:GO:0016021 | P:pigment biosynthetic process; F:catechol oxidase activity; F:tyrosinase activity; F:metal ion binding; C:integral component of membrane | Tyrosinase; Catechol oxidase | F:catechol oxidase activity | view details | ||
| JG647939.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 694 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG645833.1 | Tinospora cordifolia | peptidyl-prolyl cis-trans isomerase | 730 | P:GO:0000413; P:GO:0006457; F:GO:0003755; F:GO:0016018; C:GO:0005737 | P:protein peptidyl-prolyl isomerization; P:protein folding; F:peptidyl-prolyl cis-trans isomerase activity; F:cyclosporin A binding; C:cytoplasm | Peptidylprolyl isomerase | P:protein peptidyl-prolyl isomerization; F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG647991.1 | Tinospora cordifolia | early nodulin-93-like | 627 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG646010.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 804 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647854.1 | Tinospora cordifolia | Ras-related protein RABB1b | 738 | P:GO:0030100; F:GO:0003924; F:GO:0005525; C:GO:0005768 | P:regulation of endocytosis; F:GTPase activity; F:GTP binding; C:endosome | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG647542.1 | Tinospora cordifolia | 60S ribosomal protein L12-1 | 740 | P:GO:0006412; F:GO:0003735; F:GO:0070180; C:GO:0022625 | P:translation; F:structural constituent of ribosome; F:large ribosomal subunit rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646561.1 | Tinospora cordifolia | non-specific lipid transfer protein GPI-anchored 1-like | 719 | C:GO:0031224 | C:intrinsic component of membrane | no GO terms | view details | |||
| JG649634.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 642 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG650081.1 | Tinospora cordifolia | 7-deoxyloganetin glucosyltransferase-like | 518 | F:GO:0015020; F:GO:0080043; F:GO:0080044 | F:glucuronosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Glucuronosyltransferase | F:UDP-glycosyltransferase activity | view details | ||
| JG649358.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 811 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647703.1 | Tinospora cordifolia | Methylesterase 17 | 751 | P:GO:0009694; P:GO:0009696; F:GO:0080030; F:GO:0080031; F:GO:0080032 | P:jasmonic acid metabolic process; P:salicylic acid metabolic process; F:methyl indole-3-acetate esterase activity; F:methyl salicylate esterase activity; F:methyl jasmonate esterase activity | Carboxylesterase | no GO terms | view details | ||
| JG647157.1 | Tinospora cordifolia | putative transmembrane protein | 531 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650269.1 | Tinospora cordifolia | leucine-rich repeat protein 1-like | 769 | P:GO:0016310; F:GO:0016301; F:GO:0032440 | P:phosphorylation; F:kinase activity; F:2-alkenal reductase [NAD(P)+] activity | 2-alkenal reductase (NAD(P)(+)); Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JG648550.1 | Tinospora cordifolia | copper transport protein ATX1-like | 528 | F:GO:0046872 | F:metal ion binding | F:metal ion binding | view details | |||
| JG646304.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 803 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646897.1 | Tinospora cordifolia | 40S ribosomal protein S15 | 543 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG647922.1 | Tinospora cordifolia | thaumatin-like protein | 778 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650867.1 | Tinospora cordifolia | cis-3-alkyl-4-alkyloxetan-2-one decarboxylase | 754 | F:GO:0018786 | F:haloalkane dehalogenase activity | Haloalkane dehalogenase | no GO terms | view details | ||
| JG650263.1 | Tinospora cordifolia | Adagio protein 1 | 867 | P:GO:0009785; P:GO:0009908; P:GO:0010114; P:GO:0010498; P:GO:0016567; P:GO:0043153; F:GO:0009882; C:GO:0005634; C:GO:0005829; C:GO:0016021; C:GO:0019005 | P:blue light signaling pathway; P:flower development; P:response to red light; P:proteasomal protein catabolic process; P:protein ubiquitination; P:entrainment of circadian clock by photoperiod; F:blue light photoreceptor activity; C:nucleus; C:cytosol; C:integral component of membrane; C:SCF ubiquitin ligase complex | F:protein binding | view details | |||
| JG645879.1 | Tinospora cordifolia | mediator of RNA polymerase II transcription subunit 25-like | 887 | P:GO:0045944; C:GO:0005667; C:GO:0016592 | P:positive regulation of transcription by RNA polymerase II; C:transcription regulator complex; C:mediator complex | no GO terms | view details | |||
| JG647150.1 | Tinospora cordifolia | mediator of RNA polymerase II transcription subunit 22a-like | 682 | P:GO:0006357; F:GO:0003712; C:GO:0016592 | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | view details | |||
| KT384091.1 | Tinospora cordifolia | ---NA--- | 398 | no IPS match | view details | |||||
| JG649180.1 | Tinospora cordifolia | ripening-related protein grip22 | 768 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649711.1 | Tinospora cordifolia | kxDL motif-containing protein 1 | 569 | P:GO:0032418; C:GO:0099078 | P:lysosome localization; C:BORC complex | no GO terms | view details | |||
| JG647962.1 | Tinospora cordifolia | thaumatin-like protein | 790 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647877.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 640 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG645589.1 | Tinospora cordifolia | pathogenesis-related protein 1-like | 665 | C:GO:0005615 | C:extracellular space | no GO terms | view details | |||
| JG650921.1 | Tinospora cordifolia | L-ascorbate oxidase homolog | 817 | F:GO:0005507; F:GO:0016491 | F:copper ion binding; F:oxidoreductase activity | Oxidoreductases | F:copper ion binding; F:oxidoreductase activity | view details | ||
| JG646681.1 | Tinospora cordifolia | 60S ribosomal protein L6 | 832 | P:GO:0000027; P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:ribosomal large subunit assembly; P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650430.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 611 | F:protein binding | view details | |||||
| JG646430.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 712 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650548.1 | Tinospora cordifolia | exocyst complex component EXO70B1-like | 826 | P:GO:0006887; P:GO:0015031; C:GO:0000145 | P:exocytosis; P:protein transport; C:exocyst | P:exocytosis; F:phosphatidylinositol-4,5-bisphosphate binding; C:exocyst | view details | |||
| JG650954.1 | Tinospora cordifolia | polyadenylate-binding protein RBP45-like isoform X1 | 825 | F:GO:0003729; C:GO:0005829; C:GO:0016021 | F:mRNA binding; C:cytosol; C:integral component of membrane | F:nucleic acid binding; F:RNA binding | view details | |||
| JG650487.1 | Tinospora cordifolia | Glycerophosphodiester phosphodiesterase gde1 | 826 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG647340.1 | Tinospora cordifolia | ---NA--- | 822 | no GO terms | view details | |||||
| JG648408.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 646 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG650161.1 | Tinospora cordifolia | ---NA--- | 820 | no GO terms | view details | |||||
| JG647149.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 612 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650600.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 variant 1D | 744 | P:GO:0006301; P:GO:0070534; C:GO:0005634 | P:postreplication repair; P:protein K63-linked ubiquitination; C:nucleus | no GO terms | view details | |||
| JG648473.1 | Tinospora cordifolia | NTF2-like domain containing protein | 882 | no GO terms | view details | |||||
| JG647615.1 | Tinospora cordifolia | ---NA--- | 845 | no GO terms | view details | |||||
| JG646884.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 582 | F:protein binding | view details | |||||
| JG645523.1 | Tinospora cordifolia | ASCH domain | 803 | no GO terms | view details | |||||
| JG647120.1 | Tinospora cordifolia | protein DEHYDRATION-INDUCED 19 homolog 3-like isoform X2 | 708 | no GO terms | view details | |||||
| JG647644.1 | Tinospora cordifolia | nuclear transport factor 2B | 642 | P:GO:0006913; C:GO:0044613 | P:nucleocytoplasmic transport; C:nuclear pore central transport channel | P:nucleocytoplasmic transport | view details | |||
| JG648841.1 | Tinospora cordifolia | 40S ribosomal protein S5 | 777 | P:GO:0000028; P:GO:0006412; F:GO:0003729; F:GO:0003735; F:GO:0019843; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:mRNA binding; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:small ribosomal subunit | view details | |||
| JG646258.1 | Tinospora cordifolia | 60S acidic ribosomal protein P2-like | 454 | P:GO:0002182; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translational elongation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:cytoplasmic translational elongation; P:translational elongation; F:structural constituent of ribosome; C:ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG648336.1 | Tinospora cordifolia | 14-3-3-like protein | 814 | P:GO:0007165; P:GO:0046686; P:GO:0090378; F:GO:0005524; F:GO:0019904; F:GO:0043130; F:GO:0046982; C:GO:0005618; C:GO:0005739; C:GO:0005794; C:GO:0005829; C:GO:0005886 | P:signal transduction; P:response to cadmium ion; P:seed trichome elongation; F:ATP binding; F:protein domain specific binding; F:ubiquitin binding; F:protein heterodimerization activity; C:cell wall; C:mitochondrion; C:Golgi apparatus; C:cytosol; C:plasma membrane | no GO terms | view details | |||
| JG650090.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 767 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Lyases; Nucleoside-triphosphate phosphatase | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG648613.1 | Tinospora cordifolia | centromere protein C isoform X1 | 757 | P:GO:0006996; C:GO:0043229 | P:organelle organization; C:intracellular organelle | P:kinetochore assembly; F:centromeric DNA binding; C:kinetochore | view details | |||
| JG647524.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 27 | 786 | P:GO:0000209; P:GO:0043161; F:GO:0005524; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:proteasome-mediated ubiquitin-dependent protein catabolic process; F:ATP binding; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | F:protein binding | view details | ||
| JG646311.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 498 | F:protein binding | view details | |||||
| JG648730.1 | Tinospora cordifolia | copper transport protein ATX1-like | 530 | F:GO:0046872 | F:metal ion binding | F:metal ion binding | view details | |||
| JG646549.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 629 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG649777.1 | Tinospora cordifolia | probable acylpyruvase FAHD1, mitochondrial | 803 | F:GO:0018773; C:GO:0005739 | F:acetylpyruvate hydrolase activity; C:mitochondrion | Acetylpyruvate hydrolase | F:catalytic activity | view details | ||
| JG650341.1 | Tinospora cordifolia | thaumatin-like protein | 798 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648331.1 | Tinospora cordifolia | cytochrome c oxidase subunit 6b-1-like | 875 | C:GO:0005739; C:GO:0045277 | C:mitochondrion; C:respiratory chain complex IV | C:mitochondrion; C:respiratory chain complex IV | view details | |||
| JG647261.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 598 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG646115.1 | Tinospora cordifolia | Capsid polyprotein | 813 | no GO terms | view details | |||||
| JG650481.1 | Tinospora cordifolia | nicotinamidase 1-like | 826 | P:GO:0019365; F:GO:0008936 | P:pyridine nucleotide salvage; F:nicotinamidase activity | Nicotinamidase | P:pyridine nucleotide salvage; F:nicotinamidase activity | view details | ||
| JG647955.1 | Tinospora cordifolia | polyadenylate-binding protein RBP45-like isoform X2 | 801 | F:GO:0003729; C:GO:0005829; C:GO:0016021 | F:mRNA binding; C:cytosol; C:integral component of membrane | F:nucleic acid binding; F:RNA binding | view details | |||
| JG645768.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 592 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG649849.1 | Tinospora cordifolia | uncharacterized protein LOC122659955 | 806 | C:GO:0009706; C:GO:0016021 | C:chloroplast inner membrane; C:integral component of membrane | no GO terms | view details | |||
| JG648090.1 | Tinospora cordifolia | thaumatin-like protein | 861 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648342.1 | Tinospora cordifolia | BOI-related E3 ubiquitin-protein ligase 1-like | 801 | P:GO:0016567; F:GO:0004842; F:GO:0046872 | P:protein ubiquitination; F:ubiquitin-protein transferase activity; F:metal ion binding | no GO terms | view details | |||
| JG648212.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 596 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG649785.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 800 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649942.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 618 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649093.1 | Tinospora cordifolia | polyamine oxidase 2 | 719 | P:GO:0006598; F:GO:0046592; F:GO:0052901; F:GO:0052902; F:GO:0052903; F:GO:0052904; C:GO:0005777 | P:polyamine catabolic process; F:polyamine oxidase activity; F:spermine:oxygen oxidoreductase (spermidine-forming) activity; F:spermidine:oxygen oxidoreductase (3-aminopropanal-forming) activity; F:N1-acetylspermine:oxygen oxidoreductase (3-acetamidopropanal-forming) activity; F:N1-acetylspermidine:oxygen oxidoreductase (3-acetamidopropanal-forming) activity; C:peroxisome | Non-specific polyamine oxidase; Spermine oxidase; N(1)-acetylpolyamine oxidase | F:oxidoreductase activity | view details | ||
| JG650130.1 | Tinospora cordifolia | MOB kinase activator-like 1A | 819 | P:GO:0007165; P:GO:0032147; F:GO:0016301; F:GO:0030295; C:GO:0005634; C:GO:0005737 | P:signal transduction; P:activation of protein kinase activity; F:kinase activity; F:protein kinase activator activity; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG646387.1 | Tinospora cordifolia | calmodulin-7 | 794 | F:GO:0005509 | F:calcium ion binding | F:calcium ion binding | view details | |||
| JG647005.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 805 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650445.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 678 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649157.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 556 | F:protein binding | view details | |||||
| JG649138.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 724 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG648892.1 | Tinospora cordifolia | sterol carrier protein 2-like | 596 | C:GO:0005829 | C:cytosol | no GO terms | view details | |||
| JG649330.1 | Tinospora cordifolia | Aha1 N domain-containing protein | 784 | P:GO:0006457; P:GO:0032781; F:GO:0001671; F:GO:0051087; F:GO:0051879; C:GO:0005829 | P:protein folding; P:positive regulation of ATP-dependent activity; F:ATPase activator activity; F:chaperone binding; F:Hsp90 protein binding; C:cytosol | F:ATPase activator activity; F:chaperone binding; F:Hsp90 protein binding | view details | |||
| JG646243.1 | Tinospora cordifolia | pathogen-related protein-like | 665 | no GO terms | view details | |||||
| JG646988.1 | Tinospora cordifolia | ADP-ribosylation factor 2 isoform X1 | 793 | P:GO:0006471; P:GO:0006886; P:GO:0016192; F:GO:0003924; F:GO:0005525 | P:protein ADP-ribosylation; P:intracellular protein transport; P:vesicle-mediated transport; F:GTPase activity; F:GTP binding | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG648739.1 | Tinospora cordifolia | probable glutathione peroxidase 2 | 772 | P:GO:0006979; P:GO:0098869; F:GO:0004602; C:GO:0005829; C:GO:0016021 | P:response to oxidative stress; P:cellular oxidant detoxification; F:glutathione peroxidase activity; C:cytosol; C:integral component of membrane | Glutathione peroxidase | P:response to oxidative stress; F:glutathione peroxidase activity | view details | ||
| JG649335.1 | Tinospora cordifolia | Burp domain protein rd22 | 617 | no GO terms | view details | |||||
| JG650183.1 | Tinospora cordifolia | 40S ribosomal protein S15 | 674 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG645574.1 | Tinospora cordifolia | EKC/KEOPS complex subunit Tprkb | 739 | P:GO:0000722; P:GO:0002949; C:GO:0000408; C:GO:0005634; C:GO:0005829 | P:telomere maintenance via recombination; P:tRNA threonylcarbamoyladenosine modification; C:EKC/KEOPS complex; C:nucleus; C:cytosol | no GO terms | view details | |||
| JG646922.1 | Tinospora cordifolia | glucose-6-phosphate 1-dehydrogenase 6, cytoplasmic | 768 | P:GO:0006006; P:GO:0006511; P:GO:0007166; P:GO:0009051; F:GO:0004345; F:GO:0050661; C:GO:0005634; C:GO:0016021 | P:glucose metabolic process; P:ubiquitin-dependent protein catabolic process; P:cell surface receptor signaling pathway; P:pentose-phosphate shunt, oxidative branch; F:glucose-6-phosphate dehydrogenase activity; F:NADP binding; C:nucleus; C:integral component of membrane | Glucose-6-phosphate dehydrogenase (NADP(+)) | P:glucose metabolic process; F:oxidoreductase activity, acting on CH-OH group of donors; F:NADP binding | view details | ||
| JG647673.1 | Tinospora cordifolia | pathogenesis-related protein 1 | 723 | P:GO:0009409; P:GO:0009611; P:GO:0009617; P:GO:0009646; P:GO:0009651; P:GO:0009733; P:GO:0009735; P:GO:0009737; P:GO:0009739; P:GO:0009751; P:GO:0042542; P:GO:0046688; P:GO:0050790; P:GO:0050794; P:GO:0050832; F:GO:0003723; F:GO:0004540; C:GO:0005622 | P:response to cold; P:response to wounding; P:response to bacterium; P:response to absence of light; P:response to salt stress; P:response to auxin; P:response to cytokinin; P:response to abscisic acid; P:response to gibberellin; P:response to salicylic acid; P:response to hydrogen peroxide; P:response to copper ion; P:regulation of catalytic activity; P:regulation of cellular process; P:defense response to fungus; F:RNA binding; F:ribonuclease activity; C:intracellular anatomical structure | Acting on ester bonds | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | ||
| JG650530.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 586 | F:protein binding | view details | |||||
| JG650738.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 616 | F:protein binding | view details | |||||
| JG650862.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 668 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646364.1 | Tinospora cordifolia | UDP-glucose:glycoprotein glucosyltransferase | 736 | P:GO:0018279; P:GO:0071712; P:GO:0097359; F:GO:0003980; F:GO:0051082; C:GO:0005788; C:GO:0016021 | P:protein N-linked glycosylation via asparagine; P:ER-associated misfolded protein catabolic process; P:UDP-glucosylation; F:UDP-glucose:glycoprotein glucosyltransferase activity; F:unfolded protein binding; C:endoplasmic reticulum lumen; C:integral component of membrane | Glycosyltransferases | P:protein glycosylation; F:UDP-glucose:glycoprotein glucosyltransferase activity | view details | ||
| JG646954.1 | Tinospora cordifolia | major strawberry allergen Fra a 1.07-like | 608 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650534.1 | Tinospora cordifolia | thaumatin-like protein | 649 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650808.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 913 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG650873.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 803 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645642.1 | Tinospora cordifolia | mediator of RNA polymerase II transcription subunit 22b | 667 | P:GO:0006357; F:GO:0003712; C:GO:0016592 | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | view details | |||
| JG648136.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 770 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650313.1 | Tinospora cordifolia | non-specific phospholipase C3-like | 810 | F:GO:0016788 | F:hydrolase activity, acting on ester bonds | Acting on ester bonds | F:hydrolase activity, acting on ester bonds | view details | ||
| JG648903.1 | Tinospora cordifolia | 40S ribosomal protein S7 | 817 | P:GO:0006364; P:GO:0006412; P:GO:0042274; F:GO:0003735; C:GO:0022627; C:GO:0032040 | P:rRNA processing; P:translation; P:ribosomal small subunit biogenesis; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit; C:small-subunit processome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649546.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 808 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650880.1 | Tinospora cordifolia | 40S ribosomal protein S15 | 812 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG650479.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 812 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650187.1 | Tinospora cordifolia | NAC domain-containing protein 72-like | 813 | P:GO:0006355; F:GO:0003677 | P:regulation of DNA-templated transcription; F:DNA binding | no GO terms | view details | |||
| JG650260.1 | Tinospora cordifolia | NAC domain-containing protein 92 | 824 | P:GO:0006355; F:GO:0003677 | P:regulation of DNA-templated transcription; F:DNA binding | no IPS match | view details | |||
| JG649454.1 | Tinospora cordifolia | major strawberry allergen Fra a 1-2-like | 614 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648606.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 801 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648670.1 | Tinospora cordifolia | kunitz trypsin inhibitor 5-like | 776 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650520.1 | Tinospora cordifolia | RING-H2 finger protein ATL66-like | 795 | P:GO:0016567; C:GO:0016020; C:GO:0016021 | P:protein ubiquitination; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG649350.1 | Tinospora cordifolia | 60S ribosomal protein L35 | 617 | P:GO:0000463; P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); P:translation; F:structural constituent of ribosome; C:ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG648546.1 | Tinospora cordifolia | stigma-specific STIG1-like protein 1 | 677 | no GO terms | view details | |||||
| JG649806.1 | Tinospora cordifolia | thaumatin-like protein | 752 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650306.1 | Tinospora cordifolia | calnexin homolog | 806 | P:GO:0006457; F:GO:0005509; F:GO:0030246; F:GO:0051082; C:GO:0005783; C:GO:0016021 | P:protein folding; F:calcium ion binding; F:carbohydrate binding; F:unfolded protein binding; C:endoplasmic reticulum; C:integral component of membrane | P:protein folding; F:calcium ion binding; F:protein binding; F:unfolded protein binding; C:endoplasmic reticulum | view details | |||
| JG647184.1 | Tinospora cordifolia | calmodulin-binding protein 60 A isoform X1 | 816 | P:GO:0031326; F:GO:0005488 | P:regulation of cellular biosynthetic process; F:binding | F:calmodulin binding | view details | |||
| JG645696.1 | Tinospora cordifolia | DUF4228 domain-containing protein | 676 | P:GO:0016310; F:GO:0016301 | P:phosphorylation; F:kinase activity | no GO terms | view details | |||
| JG646488.1 | Tinospora cordifolia | Glutamyl-tRNA(Gln) amidotransferase subunit D like | 788 | F:GO:0016740; C:GO:0016021 | F:transferase activity; C:integral component of membrane | Transferases | no GO terms | view details | ||
| JG650489.1 | Tinospora cordifolia | thaumatin-like protein | 786 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649709.1 | Tinospora cordifolia | nucleoside diphosphate kinase B | 561 | P:GO:0006165; P:GO:0006183; P:GO:0006228; P:GO:0006241; F:GO:0004550; F:GO:0005524 | P:nucleoside diphosphate phosphorylation; P:GTP biosynthetic process; P:UTP biosynthetic process; P:CTP biosynthetic process; F:nucleoside diphosphate kinase activity; F:ATP binding | Nucleoside-diphosphate kinase | P:nucleoside diphosphate phosphorylation; P:GTP biosynthetic process; P:UTP biosynthetic process; P:CTP biosynthetic process; F:nucleoside diphosphate kinase activity | view details | ||
| JG648820.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 793 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647011.1 | Tinospora cordifolia | transmembrane protein 205 | 754 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG647779.1 | Tinospora cordifolia | ripening-related protein grip22 | 788 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG645627.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 743 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646195.1 | Tinospora cordifolia | thaumatin-like protein | 662 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650196.1 | Tinospora cordifolia | subtilisin-like protease SBT3.17 | 488 | P:GO:0006508; P:GO:0016310; F:GO:0008233; F:GO:0016301 | P:proteolysis; P:phosphorylation; F:peptidase activity; F:kinase activity | Acting on peptide bonds (peptidases); Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG648861.1 | Tinospora cordifolia | Proline-rich receptor-like protein kinase PERK9 | 799 | P:GO:0006468; F:GO:0004674; F:GO:0005524; C:GO:0005886; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; C:plasma membrane; C:integral component of membrane | Transferring phosphorus-containing groups | no IPS match | view details | ||
| JG645887.1 | Tinospora cordifolia | 60S ribosomal protein L23 | 629 | P:GO:0006412; F:GO:0003735; F:GO:0070180; C:GO:0022625 | P:translation; F:structural constituent of ribosome; F:large ribosomal subunit rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647649.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 617 | F:protein binding | view details | |||||
| JG646709.1 | Tinospora cordifolia | SH3 domain-containing protein 2-like | 748 | C:GO:0016020 | C:membrane | F:protein binding | view details | |||
| JG648403.1 | Tinospora cordifolia | chitinase-like protein 1 | 740 | P:GO:0000272; P:GO:0006032; P:GO:0006952; P:GO:0016998; F:GO:0004568; F:GO:0008061 | P:polysaccharide catabolic process; P:chitin catabolic process; P:defense response; P:cell wall macromolecule catabolic process; F:chitinase activity; F:chitin binding | Chitinase | P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG645623.1 | Tinospora cordifolia | ---NA--- | 845 | no GO terms | view details | |||||
| JG646591.1 | Tinospora cordifolia | Bet v I domain | 783 | F:GO:0016829; C:GO:0016020 | F:lyase activity; C:membrane | Lyases | P:defense response | view details | ||
| JG646351.1 | Tinospora cordifolia | cAMP-regulated phosphoprotein 19-related protein | 744 | P:GO:0035308; P:GO:0043086; F:GO:0004864; C:GO:0005737 | P:negative regulation of protein dephosphorylation; P:negative regulation of catalytic activity; F:protein phosphatase inhibitor activity; C:cytoplasm | no GO terms | view details | |||
| JG649212.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 714 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650343.1 | Tinospora cordifolia | ---NA--- | 663 | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||||
| JG647353.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 740 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG647958.1 | Tinospora cordifolia | transcription factor bHLH106 | 734 | F:GO:0003677; F:GO:0046983; C:GO:0005634 | F:DNA binding; F:protein dimerization activity; C:nucleus | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity; F:protein dimerization activity | view details | |||
| JG645941.1 | Tinospora cordifolia | histone H3 | 614 | F:GO:0003677; F:GO:0046982; C:GO:0000786; C:GO:0005634 | F:DNA binding; F:protein heterodimerization activity; C:nucleosome; C:nucleus | F:DNA binding; F:structural constituent of chromatin; F:protein heterodimerization activity; C:nucleosome | view details | |||
| JG648101.1 | Tinospora cordifolia | NAC transcription factor NAM-B1 | 833 | P:GO:0006355; F:GO:0003677 | P:regulation of DNA-templated transcription; F:DNA binding | P:regulation of DNA-templated transcription; F:DNA binding | view details | |||
| JG646029.1 | Tinospora cordifolia | probable folate-biopterin transporter 6 | 797 | P:GO:0006817; P:GO:0050896; C:GO:0016021 | P:phosphate ion transport; P:response to stimulus; C:integral component of membrane | no GO terms | view details | |||
| JG648286.1 | Tinospora cordifolia | 60S acidic ribosomal protein P2B | 618 | P:GO:0002182; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translational elongation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:cytoplasmic translational elongation; P:translational elongation; F:structural constituent of ribosome; C:ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG646710.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 771 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648864.1 | Tinospora cordifolia | thaumatin-like protein | 769 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648189.1 | Tinospora cordifolia | transmembrane emp24 domain-containing protein p24delta3-like | 781 | P:GO:0006886; P:GO:0006888; P:GO:0007030; C:GO:0005783; C:GO:0005793; C:GO:0005794; C:GO:0016021; C:GO:0030134 | P:intracellular protein transport; P:endoplasmic reticulum to Golgi vesicle-mediated transport; P:Golgi organization; C:endoplasmic reticulum; C:endoplasmic reticulum-Golgi intermediate compartment; C:Golgi apparatus; C:integral component of membrane; C:COPII-coated ER to Golgi transport vesicle | no GO terms | view details | |||
| JG650150.1 | Tinospora cordifolia | thaumatin-like protein | 795 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG645940.1 | Tinospora cordifolia | Glutamate/phenylalanine/leucine/valine dehydrogenase | 788 | P:GO:0006538; F:GO:0000166; F:GO:0004352; C:GO:0005739 | P:glutamate catabolic process; F:nucleotide binding; F:glutamate dehydrogenase (NAD+) activity; C:mitochondrion | Glutamate dehydrogenase (NAD(P)(+)); Glutamate dehydrogenase | P:cellular amino acid metabolic process; F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor | view details | ||
| JG647952.1 | Tinospora cordifolia | U-box domain-containing protein 35 isoform X1 | 821 | F:GO:0016787 | F:hydrolase activity | no GO terms | view details | |||
| JG646091.1 | Tinospora cordifolia | CLAVATA3/ESR (CLE)-related protein 12 | 613 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| JG649815.1 | Tinospora cordifolia | hypothetical protein BVC80_665g4 | 802 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG645596.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 675 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650549.1 | Tinospora cordifolia | nascent polypeptide-associated complex subunit beta-like | 739 | C:GO:0005829; C:GO:0005854; C:GO:0042788 | C:cytosol; C:nascent polypeptide-associated complex; C:polysomal ribosome | no GO terms | view details | |||
| JG646182.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 585 | F:protein binding | view details | |||||
| JG648245.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 757 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650801.1 | Tinospora cordifolia | kunitz trypsin inhibitor 5-like | 762 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648332.1 | Tinospora cordifolia | metallothiol transferase FosB | 848 | F:GO:0016829 | F:lyase activity | Lyases | no GO terms | view details | ||
| JG647653.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase | 786 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG650687.1 | Tinospora cordifolia | tRNA dimethylallyltransferase 2 isoform X1 | 838 | P:GO:0006400; F:GO:0005524; F:GO:0052381; C:GO:0005739; C:GO:0016021 | P:tRNA modification; F:ATP binding; F:tRNA dimethylallyltransferase activity; C:mitochondrion; C:integral component of membrane | tRNA dimethylallyltransferase | P:tRNA processing | view details | ||
| JG650781.1 | Tinospora cordifolia | thaumatin-like protein | 872 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648593.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 779 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648700.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 799 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649819.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 759 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647697.1 | Tinospora cordifolia | GTP-binding protein YPTM2 | 839 | P:GO:0006886; F:GO:0003924; F:GO:0005525; C:GO:0012505 | P:intracellular protein transport; F:GTPase activity; F:GTP binding; C:endomembrane system | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG649207.1 | Tinospora cordifolia | putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 | 775 | P:GO:0032259; P:GO:0043086; P:GO:0051252; F:GO:0008168; F:GO:0008428; F:GO:0008948; F:GO:0046872; F:GO:0047443 | P:methylation; P:negative regulation of catalytic activity; P:regulation of RNA metabolic process; F:methyltransferase activity; F:ribonuclease inhibitor activity; F:oxaloacetate decarboxylase activity; F:metal ion binding; F:4-hydroxy-4-methyl-2-oxoglutarate aldolase activity | 4-hydroxy-4-methyl-2-oxoglutarate aldolase; Oxaloacetate decarboxylase; Malate dehydrogenase (oxaloacetate-decarboxylating); Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP(+)); Transferring one-carbon groups | P:regulation of RNA metabolic process; F:ribonuclease inhibitor activity | view details | ||
| JG646437.1 | Tinospora cordifolia | Acyl carrier protein | 771 | P:GO:0006633; F:GO:0000036; F:GO:0031177; C:GO:0009507 | P:fatty acid biosynthetic process; F:acyl carrier activity; F:phosphopantetheine binding; C:chloroplast | P:fatty acid biosynthetic process; F:acyl carrier activity; F:phosphopantetheine binding | view details | |||
| JG646598.1 | Tinospora cordifolia | actin-depolymerizing factor 2 | 799 | P:GO:0030042; F:GO:0051015; C:GO:0005737; C:GO:0015629 | P:actin filament depolymerization; F:actin filament binding; C:cytoplasm; C:actin cytoskeleton | P:actin filament depolymerization; F:actin binding; C:actin cytoskeleton | view details | |||
| JG646299.1 | Tinospora cordifolia | autophagy-related protein 8f | 732 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016020 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:membrane | no GO terms | view details | |||
| JG646861.1 | Tinospora cordifolia | phosphoinositide phosphatase SAC1-like | 583 | P:GO:0036092; P:GO:0046856; F:GO:0043813; C:GO:0005774 | P:phosphatidylinositol-3-phosphate biosynthetic process; P:phosphatidylinositol dephosphorylation; F:phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity; C:vacuolar membrane | Acting on ester bonds | P:phosphatidylinositol dephosphorylation; F:phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity | view details | ||
| JG649114.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 583 | F:protein binding | view details | |||||
| MH577056.1 | Tinospora cordifolia | 150945 | F:ATP binding; F:ATP hydrolysis activity | view details | ||||||
| JG645844.1 | Tinospora cordifolia | polyphenol oxidase I, chloroplastic-like | 859 | P:GO:0046148; F:GO:0004097; F:GO:0004503; F:GO:0046872; C:GO:0016021 | P:pigment biosynthetic process; F:catechol oxidase activity; F:tyrosinase activity; F:metal ion binding; C:integral component of membrane | Tyrosinase; Catechol oxidase | F:catechol oxidase activity; F:oxidoreductase activity | view details | ||
| JG646489.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 839 | P:GO:0006508; F:GO:0004190; C:GO:0016021 | P:proteolysis; F:aspartic-type endopeptidase activity; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG650639.1 | Tinospora cordifolia | protein C2-DOMAIN ABA-RELATED 4-like | 683 | P:GO:0009738; C:GO:0005634; C:GO:0005886 | P:abscisic acid-activated signaling pathway; C:nucleus; C:plasma membrane | no GO terms | view details | |||
| JG648061.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 751 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG648843.1 | Tinospora cordifolia | 40S ribosomal protein S24-1 | 394 | P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0005829; C:GO:0005840 | P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosol; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649559.1 | Tinospora cordifolia | probable glutathione S-transferase | 746 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG650208.1 | Tinospora cordifolia | protein DMP3-like | 740 | P:GO:0010256; C:GO:0016021 | P:endomembrane system organization; C:integral component of membrane | no GO terms | view details | |||
| JG649739.1 | Tinospora cordifolia | 60S ribosomal protein L34-like | 609 | P:GO:0006412; P:GO:0042254; F:GO:0003735; C:GO:0022625 | P:translation; P:ribosome biogenesis; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646562.1 | Tinospora cordifolia | histone H3 | 633 | F:GO:0003677; F:GO:0046982; C:GO:0000786; C:GO:0005634 | F:DNA binding; F:protein heterodimerization activity; C:nucleosome; C:nucleus | F:DNA binding; F:structural constituent of chromatin; F:protein heterodimerization activity; C:nucleosome | view details | |||
| JG646711.1 | Tinospora cordifolia | 60S ribosomal protein L12-1 | 794 | P:GO:0006412; F:GO:0003735; F:GO:0070180; C:GO:0022625 | P:translation; F:structural constituent of ribosome; F:large ribosomal subunit rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648652.1 | Tinospora cordifolia | 60S ribosomal protein L24 | 714 | P:GO:0002181; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | no GO terms | view details | |||
| JG647913.1 | Tinospora cordifolia | ADP-ribosylation factor 2 isoform X1 | 722 | F:GO:0003924; F:GO:0005525 | F:GTPase activity; F:GTP binding | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG648360.1 | Tinospora cordifolia | zinc finger A20 and AN1 domain-containing stress-associated protein 5-like | 738 | P:GO:0006950; P:GO:0009628; P:GO:0016567; F:GO:0003677; F:GO:0004842; F:GO:0008270 | P:response to stress; P:response to abiotic stimulus; P:protein ubiquitination; F:DNA binding; F:ubiquitin-protein transferase activity; F:zinc ion binding | Transferases | F:DNA binding; F:zinc ion binding | view details | ||
| JG650819.1 | Tinospora cordifolia | Bet v I domain | 689 | P:GO:0006952 | P:defense response | P:defense response | view details | |||
| JG648750.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 802 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649834.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 711 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646504.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 variant 1D | 798 | P:GO:0006301; P:GO:0070534; C:GO:0005634 | P:postreplication repair; P:protein K63-linked ubiquitination; C:nucleus | no GO terms | view details | |||
| JG649211.1 | Tinospora cordifolia | Pleckstrin-likey domain-containing family M member 1 | 782 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| JG646672.1 | Tinospora cordifolia | thaumatin-like protein | 815 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648130.1 | Tinospora cordifolia | CASP-like protein 5C1 | 763 | C:GO:0005886; C:GO:0016021 | C:plasma membrane; C:integral component of membrane | no GO terms | view details | |||
| JG648096.1 | Tinospora cordifolia | major strawberry allergen Fra a 1-2-like | 708 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645988.1 | Tinospora cordifolia | Polyadenylate-binding protein like | 807 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG645763.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 796 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646873.1 | Tinospora cordifolia | Mannose-binding lectin | 606 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG645766.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 529 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | F:protein binding | view details | |||
| JG648231.1 | Tinospora cordifolia | hypothetical protein IFM89_037487 | 679 | no GO terms | view details | |||||
| JG649426.1 | Tinospora cordifolia | actin-depolymerizing factor 2 | 829 | P:GO:0030042; F:GO:0051015; C:GO:0005737; C:GO:0015629 | P:actin filament depolymerization; F:actin filament binding; C:cytoplasm; C:actin cytoskeleton | P:actin filament depolymerization; F:actin binding; C:actin cytoskeleton | view details | |||
| JG650772.1 | Tinospora cordifolia | phosphoprotein ECPP44-like | 859 | P:GO:0009414; P:GO:0009415; P:GO:0009631; P:GO:0009737; C:GO:0005829; C:GO:0016020 | P:response to water deprivation; P:response to water; P:cold acclimation; P:response to abscisic acid; C:cytosol; C:membrane | P:response to water | view details | |||
| JG646085.1 | Tinospora cordifolia | GTP-binding protein at2g22870 | 868 | F:GO:0005525; F:GO:0046872 | F:GTP binding; F:metal ion binding | F:GTP binding | view details | |||
| JG645684.1 | Tinospora cordifolia | hypothetical protein HHK36_010273 | 690 | C:GO:0016020 | C:membrane | no GO terms | view details | |||
| JG646736.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 810 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648348.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 794 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646264.1 | Tinospora cordifolia | serine/threonine-protein phosphatase 4 regulatory subunit 3-like isoform X2 | 758 | no GO terms | view details | |||||
| JG649432.1 | Tinospora cordifolia | methyltransferase N6AMT1-like | 858 | P:GO:0006479; F:GO:0003676; F:GO:0008276; F:GO:0008757; C:GO:0035657 | P:protein methylation; F:nucleic acid binding; F:protein methyltransferase activity; F:S-adenosylmethionine-dependent methyltransferase activity; C:eRF1 methyltransferase complex | Transferring one-carbon groups | F:methyltransferase activity | view details | ||
| JG650963.1 | Tinospora cordifolia | probable aspartic proteinase GIP2 | 590 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG647090.1 | Tinospora cordifolia | peptidyl-prolyl cis-trans isomerase | 720 | P:GO:0000413; P:GO:0006457; F:GO:0003755; F:GO:0016018; C:GO:0005737 | P:protein peptidyl-prolyl isomerization; P:protein folding; F:peptidyl-prolyl cis-trans isomerase activity; F:cyclosporin A binding; C:cytoplasm | Peptidylprolyl isomerase | P:protein peptidyl-prolyl isomerization; F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG647551.1 | Tinospora cordifolia | NEDD8-conjugating enzyme Ubc12 | 721 | P:GO:0045116; F:GO:0005524; F:GO:0016874; F:GO:0019788; C:GO:0005634 | P:protein neddylation; F:ATP binding; F:ligase activity; F:NEDD8 transferase activity; C:nucleus | Transferases; Ligases | no GO terms | view details | ||
| JG649801.1 | Tinospora cordifolia | non-specific lipid-transfer protein 2-like | 556 | P:GO:0006869 | P:lipid transport | P:lipid transport | view details | |||
| JG648234.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 664 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648120.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 799 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648290.1 | Tinospora cordifolia | 40S ribosomal protein S23 | 595 | P:GO:0006412; F:GO:0003735; C:GO:0016021; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:integral component of membrane; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG647255.1 | Tinospora cordifolia | Methyl-cpg-binding domain-containing protein | 807 | F:GO:0003677; C:GO:0005634 | F:DNA binding; C:nucleus | no GO terms | view details | |||
| JG648885.1 | Tinospora cordifolia | phosphopantothenoylcysteine decarboxylase | 836 | P:GO:0015937; F:GO:0004633; F:GO:0010181; C:GO:0071513 | P:coenzyme A biosynthetic process; F:phosphopantothenoylcysteine decarboxylase activity; F:FMN binding; C:phosphopantothenoylcysteine decarboxylase complex | Phosphopantothenoylcysteine decarboxylase | F:catalytic activity | view details | ||
| JG645564.1 | Tinospora cordifolia | probable calcium-binding protein CML36 | 796 | F:GO:0005509 | F:calcium ion binding | F:calcium ion binding | view details | |||
| JG646964.1 | Tinospora cordifolia | 60S ribosomal protein L18a-like protein | 580 | C:GO:0005840; C:GO:0016021 | C:ribosome; C:integral component of membrane | no GO terms | view details | |||
| JG647581.1 | Tinospora cordifolia | Ubiquitin-conjugating enzyme E2 10 | 792 | P:GO:0000209; P:GO:0006511; F:GO:0005524; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ATP binding; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG650133.1 | Tinospora cordifolia | protein SUPPRESSOR OF FRI 4 | 827 | P:GO:0006355; F:GO:0003677; F:GO:0046872; C:GO:0005634 | P:regulation of DNA-templated transcription; F:DNA binding; F:metal ion binding; C:nucleus | F:DNA binding | view details | |||
| JG650199.1 | Tinospora cordifolia | NAC domain-containing protein 68-like | 780 | P:GO:0006355; F:GO:0003677 | P:regulation of DNA-templated transcription; F:DNA binding | P:regulation of DNA-templated transcription; F:DNA binding | view details | |||
| JG646566.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 738 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647442.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2-23 kDa-like | 767 | P:GO:0000209; P:GO:0006511; F:GO:0005524; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ATP binding; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG645799.1 | Tinospora cordifolia | alpha-amylase/subtilisin inhibitor-like | 740 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647170.1 | Tinospora cordifolia | BAG family molecular chaperone regulator 7-like | 716 | P:GO:0006457; P:GO:0034605; P:GO:0034620; P:GO:0070417; F:GO:0051087; C:GO:0005783 | P:protein folding; P:cellular response to heat; P:cellular response to unfolded protein; P:cellular response to cold; F:chaperone binding; C:endoplasmic reticulum | F:chaperone binding | view details | |||
| JG647016.1 | Tinospora cordifolia | Mannose-binding lectin | 665 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG650820.1 | Tinospora cordifolia | Carbon-nitrogen hydrolase | 728 | P:GO:0006107; P:GO:0006108; P:GO:0006528; P:GO:0006541; F:GO:0016746; F:GO:0050152 | P:oxaloacetate metabolic process; P:malate metabolic process; P:asparagine metabolic process; P:glutamine metabolic process; F:acyltransferase activity; F:omega-amidase activity | Acyltransferases; Omega-amidase | P:nitrogen compound metabolic process; F:hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | view details | ||
| JG648461.1 | Tinospora cordifolia | acyl-CoA-binding domain-containing protein 4 isoform X1 | 813 | P:GO:0006869; F:GO:0000062; F:GO:0008289; C:GO:0005829 | P:lipid transport; F:fatty-acyl-CoA binding; F:lipid binding; C:cytosol | no GO terms | view details | |||
| JG646687.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 814 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648051.1 | Tinospora cordifolia | protein DMP3-like | 785 | P:GO:0010256; C:GO:0016021 | P:endomembrane system organization; C:integral component of membrane | no GO terms | view details | |||
| JG647786.1 | Tinospora cordifolia | 40S ribosomal protein S20-2 | 578 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG649135.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 571 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG645792.1 | Tinospora cordifolia | nuclear transport factor 2B | 770 | P:GO:0006913; C:GO:0044613 | P:nucleocytoplasmic transport; C:nuclear pore central transport channel | P:nucleocytoplasmic transport | view details | |||
| JG647410.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 806 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646476.1 | Tinospora cordifolia | 60S acidic ribosomal protein P2-like | 584 | P:GO:0002182; F:GO:0003735; C:GO:0016021; C:GO:0022625 | P:cytoplasmic translational elongation; F:structural constituent of ribosome; C:integral component of membrane; C:cytosolic large ribosomal subunit | P:cytoplasmic translational elongation; P:translational elongation; F:structural constituent of ribosome; C:ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG648752.1 | Tinospora cordifolia | Hypothetical predicted protein | 847 | P:GO:0009639; P:GO:0051457; C:GO:0005634; C:GO:0005737 | P:response to red or far red light; P:maintenance of protein location in nucleus; C:nucleus; C:cytoplasm | P:response to red or far red light | view details | |||
| JG648389.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein LOC104608631 | 857 | no GO terms | view details | |||||
| JG646443.1 | Tinospora cordifolia | cell division cycle and apoptosis regulator protein 1 | 715 | P:GO:0006355; F:GO:0030374; C:GO:0005634 | P:regulation of DNA-templated transcription; F:nuclear receptor coactivator activity; C:nucleus | P:regulation of DNA-templated transcription; F:calcium ion binding | view details | |||
| JG645944.1 | Tinospora cordifolia | thaumatin-like protein | 808 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649794.1 | Tinospora cordifolia | 40S ribosomal protein S15 | 643 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG647050.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 581 | F:protein binding | view details | |||||
| JG650790.1 | Tinospora cordifolia | cysteine proteinase RD21A | 847 | P:GO:0051603; F:GO:0004197; F:GO:0032440; C:GO:0005615; C:GO:0005764 | P:proteolysis involved in protein catabolic process; F:cysteine-type endopeptidase activity; F:2-alkenal reductase [NAD(P)+] activity; C:extracellular space; C:lysosome | Acting on peptide bonds (peptidases); 2-alkenal reductase (NAD(P)(+)) | P:proteolysis; F:cysteine-type peptidase activity | view details | ||
| JG650254.1 | Tinospora cordifolia | BI1-like protein | 787 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG649983.1 | Tinospora cordifolia | putative indole-3-acetic acid-amido synthetase GH3.1 | 531 | no GO terms | view details | |||||
| JG645780.1 | Tinospora cordifolia | thaumatin-like protein | 818 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648110.1 | Tinospora cordifolia | Acyl carrier protein | 732 | P:GO:0006633; F:GO:0000036; F:GO:0031177; C:GO:0009507 | P:fatty acid biosynthetic process; F:acyl carrier activity; F:phosphopantetheine binding; C:chloroplast | P:fatty acid biosynthetic process; F:acyl carrier activity; F:phosphopantetheine binding | view details | |||
| JG645917.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 793 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648395.1 | Tinospora cordifolia | autophagy-related protein 8C | 668 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016020 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:membrane | no GO terms | view details | |||
| JG648956.1 | Tinospora cordifolia | cytochrome b5 | 593 | F:GO:0004128; F:GO:0009703; F:GO:0020037; F:GO:0046872; C:GO:0016021; C:GO:0043231 | F:cytochrome-b5 reductase activity, acting on NAD(P)H; F:nitrate reductase (NADH) activity; F:heme binding; F:metal ion binding; C:integral component of membrane; C:intracellular membrane-bounded organelle | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Cytochrome-b5 reductase; Nitrate reductase | no GO terms | view details | ||
| JG648358.1 | Tinospora cordifolia | Mannose-binding lectin | 685 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG649427.1 | Tinospora cordifolia | PREDICTED: uncharacterized protein LOC18611873 | 726 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG648450.1 | Tinospora cordifolia | trafficking protein particle complex subunit 1 | 829 | P:GO:0006888; C:GO:0005783; C:GO:0005794; C:GO:0030008 | P:endoplasmic reticulum to Golgi vesicle-mediated transport; C:endoplasmic reticulum; C:Golgi apparatus; C:TRAPP complex | P:vesicle-mediated transport; C:TRAPP complex | view details | |||
| JG648170.1 | Tinospora cordifolia | ras-related protein RABD1 | 800 | P:GO:0006886; F:GO:0003924; F:GO:0005525; C:GO:0012505 | P:intracellular protein transport; F:GTPase activity; F:GTP binding; C:endomembrane system | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG647606.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 815 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650140.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 28 | 730 | P:GO:0000209; P:GO:0006511; F:GO:0005524; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ATP binding; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG648382.1 | Tinospora cordifolia | GTPase LSG1-2 | 838 | F:GO:0003924; F:GO:0005525; C:GO:0005829; C:GO:0009507 | F:GTPase activity; F:GTP binding; C:cytosol; C:chloroplast | Nucleoside-triphosphate phosphatase | F:GTPase activity | view details | ||
| JG650584.1 | Tinospora cordifolia | alpha-amylase/subtilisin inhibitor-like | 709 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650914.1 | Tinospora cordifolia | epidermis-specific secreted glycoprotein EP1-like | 688 | P:GO:0048544; F:GO:0030246; C:GO:0110165 | P:recognition of pollen; F:carbohydrate binding; C:cellular anatomical entity | no GO terms | view details | |||
| JG650657.1 | Tinospora cordifolia | E3 ubiquitin-protein ligase UPL6-like | 861 | P:GO:0000209; P:GO:0006511; F:GO:0061630 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ubiquitin protein ligase activity | Transferases | P:protein polyubiquitination; F:ubiquitin-protein transferase activity; F:ubiquitin protein ligase activity | view details | ||
| JG648470.1 | Tinospora cordifolia | reverse transcriptase family protein | 674 | P:GO:0090502; F:GO:0004523; F:GO:0016779 | P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA-DNA hybrid ribonuclease activity; F:nucleotidyltransferase activity | Acting on ester bonds; Transferring phosphorus-containing groups; Ribonuclease H | no GO terms | view details | ||
| JG647061.1 | Tinospora cordifolia | probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 | 771 | F:GO:0008753; F:GO:0010181; F:GO:0050136 | F:NADPH dehydrogenase (quinone) activity; F:FMN binding; F:NADH dehydrogenase (quinone) activity | NADPH dehydrogenase (quinone); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | F:NAD(P)H dehydrogenase (quinone) activity; F:FMN binding; F:oxidoreductase activity | view details | ||
| JG645594.1 | Tinospora cordifolia | 60S ribosomal protein L34 | 647 | P:GO:0006412; P:GO:0042254; F:GO:0003735; C:GO:0022625 | P:translation; P:ribosome biogenesis; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648630.1 | Tinospora cordifolia | acyl-coenzyme A thioesterase 13-like | 718 | F:GO:0047617 | F:acyl-CoA hydrolase activity | Acyl-CoA hydrolase | F:acyl-CoA hydrolase activity | view details | ||
| JG648440.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 803 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648963.1 | Tinospora cordifolia | arogenate dehydrogenase 2, chloroplastic-like | 611 | P:GO:0006571; F:GO:0004665; F:GO:0008977; F:GO:0033730 | P:tyrosine biosynthetic process; F:prephenate dehydrogenase (NADP+) activity; F:prephenate dehydrogenase (NAD+) activity; F:arogenate dehydrogenase (NADP+) activity | Arogenate dehydrogenase (NADP(+)); Prephenate dehydrogenase; Prephenate dehydrogenase (NADP(+)); Arogenate dehydrogenase (NAD(P)(+)) | P:tyrosine biosynthetic process; F:arogenate dehydrogenase (NADP+) activity | view details | ||
| JG647256.1 | Tinospora cordifolia | ripening-related protein grip22 | 785 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648244.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 588 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648506.1 | Tinospora cordifolia | S-norcoclaurine synthase 2-like | 701 | P:GO:0006952 | P:defense response | P:defense response | view details | |||
| JG647129.1 | Tinospora cordifolia | tumor-related protein | 809 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649130.1 | Tinospora cordifolia | thaumatin-like protein | 848 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG645901.1 | Tinospora cordifolia | glutaredoxin-C9 | 778 | F:GO:0097573 | F:glutathione oxidoreductase activity | Acting on a sulfur group of donors | F:glutathione oxidoreductase activity | view details | ||
| JG648141.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 802 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648102.1 | Tinospora cordifolia | wound-induced protein 1 | 859 | no GO terms | view details | |||||
| JG647656.1 | Tinospora cordifolia | 60S ribosomal protein L13-1 | 771 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647499.1 | Tinospora cordifolia | CASP-like protein 4C1 | 752 | F:GO:0051539; C:GO:0005886; C:GO:0016021 | F:4 iron, 4 sulfur cluster binding; C:plasma membrane; C:integral component of membrane | no GO terms | view details | |||
| JG650879.1 | Tinospora cordifolia | uncharacterized protein LOC103716477 | 808 | F:GO:0005488 | F:binding | no IPS match | view details | |||
| JG646294.1 | Tinospora cordifolia | Expression of the gene is downregulated in the presence of paraquat, putative | 585 | P:GO:0006979 | P:response to oxidative stress | no GO terms | view details | |||
| JG646871.1 | Tinospora cordifolia | autophagy-related protein 8C-like | 651 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016021 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:integral component of membrane | no GO terms | view details | |||
| JG649787.1 | Tinospora cordifolia | thaumatin-like protein | 912 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646186.1 | Tinospora cordifolia | ubiquinone biosynthesis O-methyltransferase, mitochondrial-like | 630 | P:GO:0006744; P:GO:0032259; F:GO:0004395; F:GO:0008425; F:GO:0008689; C:GO:0031314 | P:ubiquinone biosynthetic process; P:methylation; F:hexaprenyldihydroxybenzoate methyltransferase activity; F:2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity; F:3-demethylubiquinone-9 3-O-methyltransferase activity; C:extrinsic component of mitochondrial inner membrane | 3-demethylubiquinol 3-O-methyltransferase; Polyprenyldihydroxybenzoate methyltransferase | no GO terms | view details | ||
| JG649597.1 | Tinospora cordifolia | glutaredoxin-C9 | 762 | F:GO:0097573 | F:glutathione oxidoreductase activity | Acting on a sulfur group of donors | F:glutathione oxidoreductase activity | view details | ||
| JG648538.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 853 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG648665.1 | Tinospora cordifolia | extracellular ribonuclease LE-like protein | 818 | P:GO:0006401; P:GO:0090502; F:GO:0003723; F:GO:0033897; C:GO:0005576 | P:RNA catabolic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA binding; F:ribonuclease T2 activity; C:extracellular region | Acting on ester bonds; Ribonuclease T(2) | F:RNA binding; F:ribonuclease T2 activity | view details | ||
| JG649151.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 790 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646403.1 | Tinospora cordifolia | ripening-related protein grip22 | 819 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647742.1 | Tinospora cordifolia | dicarboxylate transporter 2.1, chloroplastic-like | 811 | P:GO:0055085; F:GO:0022857; C:GO:0009706; C:GO:0016021 | P:transmembrane transport; F:transmembrane transporter activity; C:chloroplast inner membrane; C:integral component of membrane | Translocases | P:transmembrane transport; F:transmembrane transporter activity; C:membrane | view details | ||
| JG649512.1 | Tinospora cordifolia | AUGMIN subunit 7 isoform X1 | 602 | F:GO:0051011; C:GO:0016021 | F:microtubule minus-end binding; C:integral component of membrane | F:microtubule minus-end binding | view details | |||
| JG647041.1 | Tinospora cordifolia | bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like isoform X2 | 797 | P:GO:0008652; P:GO:0009073; P:GO:0009423; P:GO:0009793; P:GO:0019632; F:GO:0003855; F:GO:0004764; F:GO:0050661 | P:cellular amino acid biosynthetic process; P:aromatic amino acid family biosynthetic process; P:chorismate biosynthetic process; P:embryo development ending in seed dormancy; P:shikimate metabolic process; F:3-dehydroquinate dehydratase activity; F:shikimate 3-dehydrogenase (NADP+) activity; F:NADP binding | Shikimate dehydrogenase (NADP(+)); 3-dehydroquinate dehydratase; Quinate/shikimate dehydrogenase (NAD(P)(+)) | F:3-dehydroquinate dehydratase activity; F:shikimate 3-dehydrogenase (NADP+) activity | view details | ||
| JG647171.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 807 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646601.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 657 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649053.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 719 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648478.1 | Tinospora cordifolia | 40S ribosomal protein S5 | 834 | P:GO:0000028; P:GO:0006412; F:GO:0003729; F:GO:0003735; F:GO:0019843; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:mRNA binding; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:small ribosomal subunit | view details | |||
| JG648627.1 | Tinospora cordifolia | glutathione S-transferase L3 | 859 | P:GO:0006749; P:GO:0010731; P:GO:0098869; F:GO:0004364; F:GO:0045174; C:GO:0005737 | P:glutathione metabolic process; P:protein glutathionylation; P:cellular oxidant detoxification; F:glutathione transferase activity; F:glutathione dehydrogenase (ascorbate) activity; C:cytoplasm | Glutathione transferase; Glutathione dehydrogenase (ascorbate) | F:glutathione transferase activity | view details | ||
| JG650886.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 838 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Lyases; Nucleoside-triphosphate phosphatase | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG648294.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 717 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648769.1 | Tinospora cordifolia | Anaphase-promoting complex subunit like | 798 | no GO terms | view details | |||||
| JG646807.1 | Tinospora cordifolia | Bet v I domain | 681 | P:GO:0006952 | P:defense response | P:defense response | view details | |||
| JG649327.1 | Tinospora cordifolia | CASP-like protein 1D1 | 736 | F:GO:0051539; C:GO:0005886 | F:4 iron, 4 sulfur cluster binding; C:plasma membrane | no GO terms | view details | |||
| JG647347.1 | Tinospora cordifolia | coatomer subunit zeta-1-like isoform X1 | 739 | P:GO:0006886; P:GO:0006890; P:GO:0006891; C:GO:0000139; C:GO:0030126 | P:intracellular protein transport; P:retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum; P:intra-Golgi vesicle-mediated transport; C:Golgi membrane; C:COPI vesicle coat | P:retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum; C:COPI vesicle coat | view details | |||
| JG647766.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 601 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG645710.1 | Tinospora cordifolia | Dexh-box atp-dependent rna helicase dexh6 | 747 | F:GO:0003676; F:GO:0003824 | F:nucleic acid binding; F:catalytic activity | no GO terms | view details | |||
| JG647268.1 | Tinospora cordifolia | Peptidyl-prolyl cis-trans isomerase FKBP62 | 851 | P:GO:0000413; P:GO:0061077; F:GO:0003755; C:GO:0005737 | P:protein peptidyl-prolyl isomerization; P:chaperone-mediated protein folding; F:peptidyl-prolyl cis-trans isomerase activity; C:cytoplasm | Peptidylprolyl isomerase | F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG650744.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 791 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645801.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 796 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645584.1 | Tinospora cordifolia | actin-depolymerizing factor | 793 | P:GO:0030042; F:GO:0051015; C:GO:0005737; C:GO:0015629 | P:actin filament depolymerization; F:actin filament binding; C:cytoplasm; C:actin cytoskeleton | P:actin filament depolymerization; F:actin binding; C:actin cytoskeleton | view details | |||
| JG648642.1 | Tinospora cordifolia | thaumatin-like protein | 842 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647945.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 707 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648770.1 | Tinospora cordifolia | Pentatricopeptide repeat superfamily protein | 801 | no GO terms | view details | |||||
| JG646006.1 | Tinospora cordifolia | Hypothetical predicted protein | 778 | no GO terms | view details | |||||
| JG650732.1 | Tinospora cordifolia | translocon-associated protein subunit beta | 733 | C:GO:0005789; C:GO:0016021 | C:endoplasmic reticulum membrane; C:integral component of membrane | no GO terms | view details | |||
| JG648157.1 | Tinospora cordifolia | 60S ribosomal protein L34-like | 648 | P:GO:0006412; P:GO:0042254; F:GO:0003735; C:GO:0022625 | P:translation; P:ribosome biogenesis; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649810.1 | Tinospora cordifolia | NHP2-like protein 1 | 736 | P:GO:0000398; P:GO:0000470; P:GO:0030490; F:GO:0003723; C:GO:0005730; C:GO:0005840; C:GO:0031428; C:GO:0032040; C:GO:0046540; C:GO:0071011 | P:mRNA splicing, via spliceosome; P:maturation of LSU-rRNA; P:maturation of SSU-rRNA; F:RNA binding; C:nucleolus; C:ribosome; C:box C/D RNP complex; C:small-subunit processome; C:U4/U6 x U5 tri-snRNP complex; C:precatalytic spliceosome | F:RNA binding; C:nucleolus | view details | |||
| JG650853.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 807 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647605.1 | Tinospora cordifolia | ---NA--- | 828 | no GO terms | view details | |||||
| JG648301.1 | Tinospora cordifolia | Stress up-regulated Nod 19 protein | 771 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650039.1 | Tinospora cordifolia | MOB kinase activator-like 1A | 820 | P:GO:0007165; P:GO:0032147; F:GO:0016301; F:GO:0030295; C:GO:0005634; C:GO:0005737 | P:signal transduction; P:activation of protein kinase activity; F:kinase activity; F:protein kinase activator activity; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG650577.1 | Tinospora cordifolia | elongation factor 1-gamma | 716 | P:GO:0006414; P:GO:0006749; F:GO:0003746; F:GO:0004364 | P:translational elongation; P:glutathione metabolic process; F:translation elongation factor activity; F:glutathione transferase activity | Glutathione transferase | P:translational elongation; F:translation elongation factor activity; F:glutathione transferase activity | view details | ||
| JG649499.1 | Tinospora cordifolia | probable glutathione S-transferase | 780 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG648378.1 | Tinospora cordifolia | protein METHYLENE BLUE SENSITIVITY 1-like | 776 | no GO terms | view details | |||||
| JG646474.1 | Tinospora cordifolia | Non-specific lipid-transfer protein-like 1 | 652 | C:GO:0005829 | C:cytosol | no GO terms | view details | |||
| JG648406.1 | Tinospora cordifolia | ABC transporter A family member 7-like | 607 | P:GO:0006869; P:GO:0055085; F:GO:0005319; F:GO:0005524; F:GO:0140359; C:GO:0009536; C:GO:0016021 | P:lipid transport; P:transmembrane transport; F:lipid transporter activity; F:ATP binding; F:ABC-type transporter activity; C:plastid; C:integral component of membrane | Translocases | P:transmembrane transport; F:ABC-type transporter activity; C:integral component of membrane | view details | ||
| JG649788.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 745 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646526.1 | Tinospora cordifolia | heat shock 70 kDa protein, mitochondrial-like | 830 | P:GO:0034620; P:GO:0042026; P:GO:0051085; F:GO:0005524; F:GO:0016887; F:GO:0031072; F:GO:0044183; F:GO:0051082; F:GO:0051787; C:GO:0005739 | P:cellular response to unfolded protein; P:protein refolding; P:chaperone cofactor-dependent protein refolding; F:ATP binding; F:ATP hydrolysis activity; F:heat shock protein binding; F:protein folding chaperone; F:unfolded protein binding; F:misfolded protein binding; C:mitochondrion | Nucleoside-triphosphate phosphatase | F:ATP binding; F:ATP-dependent protein folding chaperone | view details | ||
| JG646272.1 | Tinospora cordifolia | 60S ribosomal protein L18a-like | 717 | P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646144.1 | Tinospora cordifolia | mediator of RNA polymerase II transcription subunit 22b | 724 | P:GO:0006357; F:GO:0003712; C:GO:0016592 | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | P:regulation of transcription by RNA polymerase II; F:transcription coregulator activity; C:mediator complex | view details | |||
| JG650513.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 710 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648912.1 | Tinospora cordifolia | Basic 7S globulin | 774 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG649858.1 | Tinospora cordifolia | nudC domain-containing protein 2 | 778 | P:GO:0006457; F:GO:0051082; C:GO:0005737 | P:protein folding; F:unfolded protein binding; C:cytoplasm | no GO terms | view details | |||
| JG650397.1 | Tinospora cordifolia | phosphoprotein ECPP44-like | 789 | P:GO:0009414; P:GO:0009415; P:GO:0009631; P:GO:0009737; C:GO:0016020 | P:response to water deprivation; P:response to water; P:cold acclimation; P:response to abscisic acid; C:membrane | P:response to water | view details | |||
| JG649468.1 | Tinospora cordifolia | TPA_asm: hypothetical protein HUJ06_005430 | 828 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no IPS match | view details | |||
| JG646782.1 | Tinospora cordifolia | thaumatin-like protein | 814 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649691.1 | Tinospora cordifolia | SAGA-associated factor 29 homolog A-like | 764 | P:GO:0043966; F:GO:0035064; C:GO:0000124 | P:histone H3 acetylation; F:methylated histone binding; C:SAGA complex | no IPS match | view details | |||
| JG647316.1 | Tinospora cordifolia | lipid transfer-like protein vas | 671 | C:GO:0016020 | C:membrane | P:lipid transport; F:lipid binding | view details | |||
| JG649852.1 | Tinospora cordifolia | thaumatin-like protein | 750 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650447.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 728 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG646090.1 | Tinospora cordifolia | 60S acidic ribosomal protein P3-like | 574 | P:GO:0006414; F:GO:0003735; C:GO:0022626 | P:translational elongation; F:structural constituent of ribosome; C:cytosolic ribosome | F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650243.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 720 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646395.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 768 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650900.1 | Tinospora cordifolia | peptidyl-prolyl cis-trans isomerase FKBP15-1-like | 780 | P:GO:0000413; P:GO:0061077; F:GO:0003755; C:GO:0016021 | P:protein peptidyl-prolyl isomerization; P:chaperone-mediated protein folding; F:peptidyl-prolyl cis-trans isomerase activity; C:integral component of membrane | Peptidylprolyl isomerase | P:protein peptidyl-prolyl isomerization; P:chaperone-mediated protein folding; F:peptidyl-prolyl cis-trans isomerase activity | view details | ||
| JG649110.1 | Tinospora cordifolia | endoglucanase 12 | 749 | P:GO:0030245; F:GO:0008810; C:GO:0016021 | P:cellulose catabolic process; F:cellulase activity; C:integral component of membrane | Cellulase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647986.1 | Tinospora cordifolia | ribonuclease 3 | 779 | P:GO:0006401; P:GO:0090502; F:GO:0003723; F:GO:0033897; C:GO:0005576 | P:RNA catabolic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA binding; F:ribonuclease T2 activity; C:extracellular region | Acting on ester bonds; Ribonuclease T(2) | F:RNA binding; F:ribonuclease T2 activity | view details | ||
| JG648013.1 | Tinospora cordifolia | universal stress protein PHOS32 | 813 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| JG649429.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2-17 kDa | 832 | P:GO:0000209; P:GO:0006511; F:GO:0005524; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ATP binding; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG646173.1 | Tinospora cordifolia | putative ribonuclease H protein | 625 | P:GO:0010027; P:GO:0010196; P:GO:0045893; P:GO:0090391; C:GO:0009515; C:GO:0016021 | P:thylakoid membrane organization; P:nonphotochemical quenching; P:positive regulation of DNA-templated transcription; P:granum assembly; C:granal stacked thylakoid; C:integral component of membrane | no GO terms | view details | |||
| JG647834.1 | Tinospora cordifolia | autophagy-related protein 8i | 646 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016020 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:membrane | no GO terms | view details | |||
| JG645579.1 | Tinospora cordifolia | Ras-related protein RABB1b | 751 | P:GO:0030100; F:GO:0003924; F:GO:0005525; C:GO:0005768 | P:regulation of endocytosis; F:GTPase activity; F:GTP binding; C:endosome | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG649558.1 | Tinospora cordifolia | protein CREG1 | 810 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649798.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 664 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645959.1 | Tinospora cordifolia | ---NA--- | 819 | no GO terms | view details | |||||
| JG649148.1 | Tinospora cordifolia | ras-related protein RABD1 | 726 | P:GO:0006886; P:GO:0006888; F:GO:0003924; F:GO:0005525; F:GO:0030742; F:GO:0080115; C:GO:0000139; C:GO:0032588 | P:intracellular protein transport; P:endoplasmic reticulum to Golgi vesicle-mediated transport; F:GTPase activity; F:GTP binding; F:GTP-dependent protein binding; F:myosin XI tail binding; C:Golgi membrane; C:trans-Golgi network membrane | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG649989.1 | Tinospora cordifolia | 7-deoxyloganetin glucosyltransferase-like | 512 | F:GO:0015020; F:GO:0080043; F:GO:0080044 | F:glucuronosyltransferase activity; F:quercetin 3-O-glucosyltransferase activity; F:quercetin 7-O-glucosyltransferase activity | Glucuronosyltransferase | F:UDP-glycosyltransferase activity | view details | ||
| JG650435.1 | Tinospora cordifolia | V-type proton ATPase subunit G 1-like | 778 | P:GO:1902600; F:GO:0042626; C:GO:0016471 | P:proton transmembrane transport; F:ATPase-coupled transmembrane transporter activity; C:vacuolar proton-transporting V-type ATPase complex | Translocases | P:proton transmembrane transport; F:proton-transporting ATPase activity, rotational mechanism; C:vacuolar proton-transporting V-type ATPase complex | view details | ||
| JG646888.1 | Tinospora cordifolia | flowering-promoting factor 1-like protein 3 | 627 | P:GO:0009909 | P:regulation of flower development | P:regulation of flower development | view details | |||
| JG648515.1 | Tinospora cordifolia | cyclin-dependent kinase inhibitor 1-like | 780 | P:GO:0051726; F:GO:0004860; C:GO:0005634 | P:regulation of cell cycle; F:protein kinase inhibitor activity; C:nucleus | P:negative regulation of cyclin-dependent protein serine/threonine kinase activity; P:regulation of cell cycle; F:cyclin-dependent protein serine/threonine kinase inhibitor activity; C:nucleus | view details | |||
| JG647920.1 | Tinospora cordifolia | succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial-like | 810 | P:GO:0006099; P:GO:0006124; P:GO:0022904; F:GO:0008177; F:GO:0009055; F:GO:0046872; F:GO:0051537; F:GO:0051538; F:GO:0051539; C:GO:0005743; C:GO:0045273 | P:tricarboxylic acid cycle; P:ferredoxin metabolic process; P:respiratory electron transport chain; F:succinate dehydrogenase (ubiquinone) activity; F:electron transfer activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; F:3 iron, 4 sulfur cluster binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrial inner membrane; C:respiratory chain complex II | Succinate dehydrogenase (quinone) | P:tricarboxylic acid cycle; F:electron transfer activity; F:oxidoreductase activity; F:iron-sulfur cluster binding | view details | ||
| JG651013.1 | Tinospora cordifolia | 60S ribosomal protein L17-2 | 726 | P:GO:0002181; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:large ribosomal subunit | view details | |||
| JG649175.1 | Tinospora cordifolia | N-alpha-acetyltransferase MAK3 | 717 | P:GO:0017196; F:GO:0004596; C:GO:0031417 | P:N-terminal peptidyl-methionine acetylation; F:peptide alpha-N-acetyltransferase activity; C:NatC complex | Acyltransferases | P:N-terminal peptidyl-methionine acetylation; F:peptide alpha-N-acetyltransferase activity; F:acetyltransferase activity | view details | ||
| JG648129.1 | Tinospora cordifolia | type II peroxiredoxin C | 716 | P:GO:0034599; P:GO:0042744; P:GO:0045454; P:GO:0098869; F:GO:0008379; C:GO:0005737 | P:cellular response to oxidative stress; P:hydrogen peroxide catabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:thioredoxin peroxidase activity; C:cytoplasm | Acting on a peroxide as acceptor | F:oxidoreductase activity | view details | ||
| JG645762.1 | Tinospora cordifolia | Exocyst complex component sec5a | 780 | P:GO:0006887; P:GO:0006893; P:GO:0015031; C:GO:0000145 | P:exocytosis; P:Golgi to plasma membrane transport; P:protein transport; C:exocyst | P:Golgi to plasma membrane transport; C:exocyst | view details | |||
| JG649868.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 802 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646288.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 694 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG649081.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP)-like | 803 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG649355.1 | Tinospora cordifolia | pre-mRNA cleavage factor Im 25 kDa subunit 2 | 781 | P:GO:0006364; P:GO:0006378; F:GO:0003729; C:GO:0005829; C:GO:0005849 | P:rRNA processing; P:mRNA polyadenylation; F:mRNA binding; C:cytosol; C:mRNA cleavage factor complex | P:mRNA polyadenylation; F:mRNA binding; C:mRNA cleavage factor complex | view details | |||
| JG647805.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 805 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650070.1 | Tinospora cordifolia | DNA repair protein | 728 | no GO terms | view details | |||||
| JG648948.1 | Tinospora cordifolia | VAMP-like protein YKT61 | 791 | P:GO:0016192; C:GO:0016021; C:GO:0043231 | P:vesicle-mediated transport; C:integral component of membrane; C:intracellular membrane-bounded organelle | no GO terms | view details | |||
| JG649189.1 | Tinospora cordifolia | 2-oxoglutarate-dependent dioxygenase DAO | 858 | P:GO:0008152; P:GO:0065008; F:GO:0051213 | P:metabolic process; P:regulation of biological quality; F:dioxygenase activity | Oxidoreductases | no GO terms | view details | ||
| JG648836.1 | Tinospora cordifolia | pathogenesis-related protein 1-like | 540 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647271.1 | Tinospora cordifolia | ripening-related protein grip22 | 794 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650042.1 | Tinospora cordifolia | protein SUPPRESSOR OF FRI 4 | 780 | P:GO:0006355; F:GO:0003677; F:GO:0046872; C:GO:0005634 | P:regulation of DNA-templated transcription; F:DNA binding; F:metal ion binding; C:nucleus | F:DNA binding | view details | |||
| JG649334.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 804 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645781.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 824 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG649714.1 | Tinospora cordifolia | NAC domain-containing protein 100 | 593 | P:GO:0006355; F:GO:0003677 | P:regulation of DNA-templated transcription; F:DNA binding | P:regulation of DNA-templated transcription; F:DNA binding | view details | |||
| JG647054.1 | Tinospora cordifolia | ADP-ribosylation factor 1-like 2 | 842 | P:GO:0006886; P:GO:0016192; F:GO:0003924; F:GO:0005525; C:GO:0009507 | P:intracellular protein transport; P:vesicle-mediated transport; F:GTPase activity; F:GTP binding; C:chloroplast | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG650104.1 | Tinospora cordifolia | Basic 7S globulin | 739 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG648571.1 | Tinospora cordifolia | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | 779 | P:GO:0006120; P:GO:0032981; F:GO:0003954; F:GO:0046872; F:GO:0051539; C:GO:0005747 | P:mitochondrial electron transport, NADH to ubiquinone; P:mitochondrial respiratory chain complex I assembly; F:NADH dehydrogenase activity; F:metal ion binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrial respiratory chain complex I | Acting on NADH or NADPH | F:oxidoreductase activity, acting on NAD(P)H; F:4 iron, 4 sulfur cluster binding; C:membrane | view details | ||
| JG647099.1 | Tinospora cordifolia | 60S ribosomal protein L11 | 779 | P:GO:0006412; F:GO:0003735; F:GO:0019843; C:GO:0005634; C:GO:0022625 | P:translation; F:structural constituent of ribosome; F:rRNA binding; C:nucleus; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646578.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 771 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650753.1 | Tinospora cordifolia | GDSL esterase/lipase At5g03610-like | 874 | F:GO:0016788 | F:hydrolase activity, acting on ester bonds | Acting on ester bonds | F:hydrolase activity, acting on ester bonds | view details | ||
| JG648426.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 803 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648793.1 | Tinospora cordifolia | prostaglandin reductase-3 | 741 | F:GO:0008270; F:GO:0016491 | F:zinc ion binding; F:oxidoreductase activity | Oxidoreductases | no GO terms | view details | ||
| JG645591.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 4 | 719 | P:GO:0000209; P:GO:0006511; F:GO:0005524; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ATP binding; F:ubiquitin conjugating enzyme activity; C:nucleus | E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG648438.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 553 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG646241.1 | Tinospora cordifolia | NADH:ubiquinone oxidoreductase | 614 | no GO terms | view details | |||||
| JG649206.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 793 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| MS688052.1 | Tinospora cordifolia | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | 1467 | P:GO:0032259; F:GO:0030776; F:GO:0030794; C:GO:0005737 | P:methylation; F:(RS)-1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase activity; F:(S)-coclaurine-N-methyltransferase activity; C:cytoplasm | (S)-coclaurine-N-methyltransferase; (RS)-1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase | no GO terms | view details | ||
| JG646149.1 | Tinospora cordifolia | nascent polypeptide-associated complex subunit beta-like | 672 | C:GO:0005829; C:GO:0005854; C:GO:0042788 | C:cytosol; C:nascent polypeptide-associated complex; C:polysomal ribosome | no GO terms | view details | |||
| JG645744.1 | Tinospora cordifolia | tryptamine hydroxycinnamoyltransferase 2-like | 729 | F:GO:0016747 | F:acyltransferase activity, transferring groups other than amino-acyl groups | Acyltransferases | no GO terms | view details | ||
| JG645987.1 | Tinospora cordifolia | kunitz trypsin inhibitor 5-like | 798 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648658.1 | Tinospora cordifolia | Kunitz trypsin inhibitor | 801 | P:GO:0006508; P:GO:0010951; F:GO:0004866; F:GO:0008233 | P:proteolysis; P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity; F:peptidase activity | F:endopeptidase inhibitor activity | view details | |||
| JG650384.1 | Tinospora cordifolia | flowering time control protein FPA | 872 | P:GO:0006378; P:GO:0009553; P:GO:0009911; P:GO:0010228; P:GO:0031048; F:GO:0003676; F:GO:0003723; C:GO:0000785; C:GO:0110165 | P:mRNA polyadenylation; P:embryo sac development; P:positive regulation of flower development; P:vegetative to reproductive phase transition of meristem; P:heterochromatin assembly by small RNA; F:nucleic acid binding; F:RNA binding; C:chromatin; C:cellular anatomical entity | no GO terms | view details | |||
| JG649498.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 760 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646511.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 796 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650785.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 682 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646417.1 | Tinospora cordifolia | thaumatin-like protein | 830 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646608.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 669 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646127.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 693 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG646180.1 | Tinospora cordifolia | thaumatin-like protein | 834 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG649828.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 530 | F:protein binding | view details | |||||
| JG647585.1 | Tinospora cordifolia | early nodulin-93-like | 723 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG645888.1 | Tinospora cordifolia | ---NA--- | 796 | no GO terms | view details | |||||
| JG646109.1 | Tinospora cordifolia | multiple organellar RNA editing factor 2, chloroplastic-like | 861 | P:GO:0016554; P:GO:0080156; C:GO:0005739 | P:cytidine to uridine editing; P:mitochondrial mRNA modification; C:mitochondrion | P:cytidine to uridine editing | view details | |||
| JG650934.1 | Tinospora cordifolia | Bet v I domain | 800 | F:GO:0016829; C:GO:0016020 | F:lyase activity; C:membrane | Lyases | P:defense response | view details | ||
| JG645945.1 | Tinospora cordifolia | pathogenesis-related protein STH-2-like | 859 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645681.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 795 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Lyases; Nucleoside-triphosphate phosphatase | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG649486.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 616 | F:protein binding | view details | |||||
| JG646501.1 | Tinospora cordifolia | temperature-induced lipocalin-1 | 800 | P:GO:0000302; P:GO:0006629; C:GO:0005737 | P:response to reactive oxygen species; P:lipid metabolic process; C:cytoplasm | no GO terms | view details | |||
| JG649153.1 | Tinospora cordifolia | thaumatin-like protein | 758 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646069.1 | Tinospora cordifolia | Glutathione S-transferase DHAR2 | 841 | P:GO:0010731; P:GO:0033355; P:GO:0098869; F:GO:0004364; F:GO:0045174 | P:protein glutathionylation; P:ascorbate glutathione cycle; P:cellular oxidant detoxification; F:glutathione transferase activity; F:glutathione dehydrogenase (ascorbate) activity | Glutathione transferase; Glutathione dehydrogenase (ascorbate) | P:ascorbate glutathione cycle; P:cellular oxidant detoxification; F:glutathione dehydrogenase (ascorbate) activity | view details | ||
| JG650810.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 819 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG650843.1 | Tinospora cordifolia | uncharacterized protein At4g13200, chloroplastic isoform X2 | 713 | P:GO:0010027; C:GO:0009535 | P:thylakoid membrane organization; C:chloroplast thylakoid membrane | P:thylakoid membrane organization | view details | |||
| JG647549.1 | Tinospora cordifolia | leucine-rich repeat protein 1-like | 810 | P:GO:0016310; F:GO:0016301; F:GO:0032440 | P:phosphorylation; F:kinase activity; F:2-alkenal reductase [NAD(P)+] activity | 2-alkenal reductase (NAD(P)(+)); Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JG648176.1 | Tinospora cordifolia | NAC domain-containing protein 83-like | 592 | P:GO:0006355; F:GO:0003677 | P:regulation of DNA-templated transcription; F:DNA binding | P:regulation of DNA-templated transcription; F:DNA binding | view details | |||
| JG650094.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 806 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650328.1 | Tinospora cordifolia | 60S ribosomal protein L22-2-like | 633 | P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0005737; C:GO:0005840 | P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytoplasm; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG646696.1 | Tinospora cordifolia | ripening-related protein grip22 | 759 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647371.1 | Tinospora cordifolia | sulfate transporter 3.1-like | 683 | P:GO:1902358; F:GO:0008271; F:GO:0015293; C:GO:0005887; C:GO:0009507 | P:sulfate transmembrane transport; F:secondary active sulfate transmembrane transporter activity; F:symporter activity; C:integral component of plasma membrane; C:chloroplast | Translocases | P:sulfate transport; P:transmembrane transport; F:sulfate transmembrane transporter activity; F:obsolete anion:anion antiporter activity; C:chloroplast; C:membrane | view details | ||
| JG646018.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 701 | F:protein binding | view details | |||||
| JG647139.1 | Tinospora cordifolia | glutamine-dependent NAD(+) synthetase | 678 | P:GO:0009435; F:GO:0003952; F:GO:0004359; F:GO:0005524; C:GO:0005737 | P:NAD biosynthetic process; F:NAD+ synthase (glutamine-hydrolyzing) activity; F:glutaminase activity; F:ATP binding; C:cytoplasm | Glutaminase; NAD(+) synthase (glutamine-hydrolyzing) | P:NAD biosynthetic process; F:NAD+ synthase (glutamine-hydrolyzing) activity; F:glutaminase activity; C:cytoplasm | view details | ||
| JG650913.1 | Tinospora cordifolia | ---NA--- | 744 | no GO terms | view details | |||||
| JG649447.1 | Tinospora cordifolia | Mannose-binding lectin | 710 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG647755.1 | Tinospora cordifolia | Glutathione S-transferase | 798 | P:GO:0006749; F:GO:0004364 | P:glutathione metabolic process; F:glutathione transferase activity | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG646467.1 | Tinospora cordifolia | lipid transfer-like protein vas | 738 | C:GO:0016020 | C:membrane | P:lipid transport; F:lipid binding | view details | |||
| JG648922.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 762 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG647500.1 | Tinospora cordifolia | probable signal peptidase complex subunit 2 | 781 | P:GO:0006465; P:GO:0045047; F:GO:0008233; C:GO:0005787; C:GO:0016021 | P:signal peptide processing; P:protein targeting to ER; F:peptidase activity; C:signal peptidase complex; C:integral component of membrane | Acting on peptide bonds (peptidases) | P:signal peptide processing; C:signal peptidase complex; C:integral component of membrane | view details | ||
| JG648401.1 | Tinospora cordifolia | ripening-related protein grip22 | 745 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648719.1 | Tinospora cordifolia | ripening-related protein grip22 | 825 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647343.1 | Tinospora cordifolia | protein enabled | 808 | no GO terms | view details | |||||
| JG648305.1 | Tinospora cordifolia | cinnamoyl-CoA reductase 1 | 738 | F:GO:0016616 | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | Acting on the CH-OH group of donors | no GO terms | view details | ||
| JG646058.1 | Tinospora cordifolia | PR5-like receptor kinase | 579 | P:GO:0006468; F:GO:0004674; F:GO:0005524; F:GO:0030247; C:GO:0016021 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding; F:polysaccharide binding; C:integral component of membrane | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:protein tyrosine kinase activity; F:ATP binding | view details | ||
| JG650283.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 703 | P:GO:0005975; F:GO:0016765; F:GO:0042973 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650276.1 | Tinospora cordifolia | probable glutathione S-transferase | 815 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG646434.1 | Tinospora cordifolia | homeobox-leucine zipper protein HOX3 | 792 | P:GO:0006357; F:GO:0000981; F:GO:0043565; C:GO:0005634 | P:regulation of transcription by RNA polymerase II; F:DNA-binding transcription factor activity, RNA polymerase II-specific; F:sequence-specific DNA binding; C:nucleus | P:regulation of DNA-templated transcription; F:DNA binding; F:sequence-specific DNA binding | view details | |||
| JG648484.1 | Tinospora cordifolia | 40S ribosomal protein S18 | 695 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0005829; C:GO:0015935 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosol; C:small ribosomal subunit | P:translation; F:nucleic acid binding; F:RNA binding; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650110.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, acidic-like | 560 | P:GO:0005975; F:GO:0016765; F:GO:0042973 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| KT384147.1 | Tinospora cordifolia | ---NA--- | 485 | no IPS match | view details | |||||
| JG646833.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 611 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646614.1 | Tinospora cordifolia | thaumatin-like protein | 826 | no GO terms | view details | |||||
| JG648816.1 | Tinospora cordifolia | CCR4-NOT transcription complex subunit 11-like | 772 | P:GO:0006402; P:GO:0017148; P:GO:0031047; C:GO:0005634; C:GO:0005737; C:GO:0030014 | P:mRNA catabolic process; P:negative regulation of translation; P:gene silencing by RNA; C:nucleus; C:cytoplasm; C:CCR4-NOT complex | C:CCR4-NOT complex | view details | |||
| JG648788.1 | Tinospora cordifolia | polyphenol oxidase, chloroplastic-like | 827 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | F:oxidoreductase activity | view details | ||
| JG648390.1 | Tinospora cordifolia | chromatin remodeling protein EBS-like | 795 | P:GO:0006325; P:GO:0006355; P:GO:0035067; P:GO:2000028; F:GO:0000976; F:GO:0003682; F:GO:0035064; F:GO:0046872; C:GO:0005634; C:GO:0016021 | P:chromatin organization; P:regulation of DNA-templated transcription; P:negative regulation of histone acetylation; P:regulation of photoperiodism, flowering; F:transcription cis-regulatory region binding; F:chromatin binding; F:methylated histone binding; F:metal ion binding; C:nucleus; C:integral component of membrane | P:chromatin organization; F:chromatin binding; F:methylated histone binding | view details | |||
| JG648555.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 638 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG646082.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2-17 kDa | 800 | P:GO:0000209; P:GO:0006511; F:GO:0004839; F:GO:0005524; F:GO:0016746; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ubiquitin activating enzyme activity; F:ATP binding; F:acyltransferase activity; F:ubiquitin conjugating enzyme activity; C:nucleus | E1 ubiquitin-activating enzyme; E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG648155.1 | Tinospora cordifolia | hypothetical protein BVC80_9031g41 | 821 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no IPS match | view details | |||
| JG648214.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 652 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649193.1 | Tinospora cordifolia | probable isoaspartyl peptidase/L-asparaginase 2 | 619 | P:GO:0016540; F:GO:0004067; F:GO:0008798; C:GO:0005737 | P:protein autoprocessing; F:asparaginase activity; F:beta-aspartyl-peptidase activity; C:cytoplasm | Asparaginase; Beta-aspartyl-peptidase | F:hydrolase activity | view details | ||
| JG650833.1 | Tinospora cordifolia | pollen-specific protein C13-like | 669 | C:GO:0005615 | C:extracellular space | no GO terms | view details | |||
| JG646429.1 | Tinospora cordifolia | protein CREG1-like | 763 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650978.1 | Tinospora cordifolia | LOC110425786 isoform | 823 | no GO terms | view details | |||||
| JG646360.1 | Tinospora cordifolia | thaumatin-like protein | 749 | no GO terms | view details | |||||
| JG646830.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 700 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646092.1 | Tinospora cordifolia | pathogenesis-related protein 1-like | 649 | C:GO:0005615 | C:extracellular space | no GO terms | view details | |||
| JG647211.1 | Tinospora cordifolia | ---NA--- | 624 | no IPS match | view details | |||||
| JG648086.1 | Tinospora cordifolia | 60S ribosomal protein L18-2 | 671 | P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649171.1 | Tinospora cordifolia | phosphoenolpyruvate carboxykinase (ATP) | 737 | P:GO:0006094; P:GO:0016310; F:GO:0004612; F:GO:0005524; F:GO:0016301; C:GO:0005829 | P:gluconeogenesis; P:phosphorylation; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:kinase activity; C:cytosol | Phosphoenolpyruvate carboxykinase (ATP); Transferring phosphorus-containing groups; Phosphoenolpyruvate carboxykinase (GTP) | P:gluconeogenesis; F:phosphoenolpyruvate carboxykinase activity; F:phosphoenolpyruvate carboxykinase (ATP) activity; F:ATP binding; F:purine nucleotide binding | view details | ||
| JG650492.1 | Tinospora cordifolia | autophagy-related protein 8C-like | 555 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016021 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:integral component of membrane | no GO terms | view details | |||
| JG648464.1 | Tinospora cordifolia | Deoxymugineic acid synthase 1 | 622 | F:GO:0004032; F:GO:0047834; C:GO:0005829 | F:alditol:NADP+ 1-oxidoreductase activity; F:D-threo-aldose 1-dehydrogenase activity; C:cytosol | Aldose reductase; D-threo-aldose 1-dehydrogenase | F:oxidoreductase activity | view details | ||
| JG648664.1 | Tinospora cordifolia | glutamyl-tRNA reductase 1, chloroplastic-like | 875 | P:GO:0006782; F:GO:0008883; F:GO:0050661 | P:protoporphyrinogen IX biosynthetic process; F:glutamyl-tRNA reductase activity; F:NADP binding | Glutamyl-tRNA reductase | P:tetrapyrrole biosynthetic process; F:glutamyl-tRNA reductase activity; F:NADP binding | view details | ||
| JG649642.1 | Tinospora cordifolia | AP-1 complex subunit sigma-2 | 637 | P:GO:0006886; P:GO:0016192; F:GO:0035615; C:GO:0030121 | P:intracellular protein transport; P:vesicle-mediated transport; F:clathrin adaptor activity; C:AP-1 adaptor complex | P:protein transport; P:vesicle-mediated transport; F:clathrin adaptor activity; C:AP-1 adaptor complex | view details | |||
| JG646341.1 | Tinospora cordifolia | formate dehydrogenase, mitochondrial | 717 | P:GO:0042183; F:GO:0008863; F:GO:0016616; F:GO:0051287; C:GO:0005739; C:GO:0009326; C:GO:0009507 | P:formate catabolic process; F:formate dehydrogenase (NAD+) activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding; C:mitochondrion; C:formate dehydrogenase complex; C:chloroplast | Acting on the CH-OH group of donors; Formate dehydrogenase; Acting on the aldehyde or oxo group of donors | F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:NAD binding | view details | ||
| JG648790.1 | Tinospora cordifolia | kunitz trypsin inhibitor 2 | 568 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649342.1 | Tinospora cordifolia | 60S ribosomal protein L18-2 | 700 | P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649684.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 799 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649754.1 | Tinospora cordifolia | Reticuline oxidase | 611 | F:GO:0016491; F:GO:0071949 | F:oxidoreductase activity; F:FAD binding | Oxidoreductases | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | ||
| JG645996.1 | Tinospora cordifolia | V-type proton ATPase subunit G 1-like | 628 | P:GO:1902600; F:GO:0042626; C:GO:0016471 | P:proton transmembrane transport; F:ATPase-coupled transmembrane transporter activity; C:vacuolar proton-transporting V-type ATPase complex | Translocases | P:proton transmembrane transport; F:proton-transporting ATPase activity, rotational mechanism; C:vacuolar proton-transporting V-type ATPase complex | view details | ||
| JG646000.1 | Tinospora cordifolia | protein Brevis radix-like 4 | 811 | C:GO:0005634 | C:nucleus | no GO terms | view details | |||
| JG648742.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase | 813 | P:GO:0002238; P:GO:0009805; F:GO:0016706; F:GO:0031418; F:GO:0046872 | P:response to molecule of fungal origin; P:coumarin biosynthetic process; F:2-oxoglutarate-dependent dioxygenase activity; F:L-ascorbic acid binding; F:metal ion binding | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no GO terms | view details | ||
| JG648588.1 | Tinospora cordifolia | CASP-like protein 1D1 | 814 | F:GO:0051539; C:GO:0005886 | F:4 iron, 4 sulfur cluster binding; C:plasma membrane | no GO terms | view details | |||
| JG650013.1 | Tinospora cordifolia | Basic 7S globulin | 729 | P:GO:0006508; F:GO:0004190 | P:proteolysis; F:aspartic-type endopeptidase activity | Acting on peptide bonds (peptidases) | P:proteolysis; F:aspartic-type endopeptidase activity | view details | ||
| JG649372.1 | Tinospora cordifolia | ATP synthase subunit d, mitochondrial | 742 | P:GO:0006811; P:GO:0015986; F:GO:0015078; C:GO:0000274 | P:ion transport; P:proton motive force-driven ATP synthesis; F:proton transmembrane transporter activity; C:mitochondrial proton-transporting ATP synthase, stator stalk | Translocases | P:proton motive force-driven ATP synthesis; F:proton transmembrane transporter activity; C:mitochondrial proton-transporting ATP synthase complex, coupling factor F(o) | view details | ||
| JG645823.1 | Tinospora cordifolia | pathogenesis-related protein 1-like | 675 | P:GO:0009607; C:GO:0005576 | P:response to biotic stimulus; C:extracellular region | no GO terms | view details | |||
| JG646048.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 670 | P:GO:0005975; F:GO:0016765; F:GO:0042973 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG649669.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 746 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG650802.1 | Tinospora cordifolia | Berberine bridge enzyme-like | 858 | F:GO:0050660 | F:flavin adenine dinucleotide binding | F:oxidoreductase activity; F:flavin adenine dinucleotide binding | view details | |||
| JG647349.1 | Tinospora cordifolia | 60S ribosomal protein L15 | 758 | P:GO:0002181; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:cytoplasmic translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649169.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 675 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647025.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 790 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646034.1 | Tinospora cordifolia | ripening-related protein grip22 | 800 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646767.1 | Tinospora cordifolia | probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 | 767 | F:GO:0010296; C:GO:0000139; C:GO:0005789; C:GO:0016021 | F:prenylcysteine methylesterase activity; C:Golgi membrane; C:endoplasmic reticulum membrane; C:integral component of membrane | Carboxylesterase | no GO terms | view details | ||
| JG647001.1 | Tinospora cordifolia | nuclear transport factor 2-like | 867 | F:GO:0003729; C:GO:0005829; C:GO:0043229; C:GO:1990904 | F:mRNA binding; C:cytosol; C:intracellular organelle; C:ribonucleoprotein complex | no GO terms | view details | |||
| JG647072.1 | Tinospora cordifolia | polyubiquitin 11 | 782 | P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0016874; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005737 | P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:ligase activity; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:cytoplasm | Ligases | F:protein binding | view details | ||
| JG645972.1 | Tinospora cordifolia | Glycoside hydrolase family 38 | 757 | P:GO:0006013; F:GO:0004559; F:GO:0030246; F:GO:0046872; C:GO:0016021 | P:mannose metabolic process; F:alpha-mannosidase activity; F:carbohydrate binding; F:metal ion binding; C:integral component of membrane | Alpha-mannosidase | P:carbohydrate metabolic process; F:catalytic activity; F:carbohydrate binding | view details | ||
| JG648281.1 | Tinospora cordifolia | kinesin-like protein KIN-13A | 771 | P:GO:0007018; F:GO:0003777; F:GO:0005524; F:GO:0008017; F:GO:0016787; C:GO:0005874; C:GO:0009507 | P:microtubule-based movement; F:microtubule motor activity; F:ATP binding; F:microtubule binding; F:hydrolase activity; C:microtubule; C:chloroplast | Hydrolases | no GO terms | view details | ||
| JG648599.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 789 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650697.1 | Tinospora cordifolia | protein CREG1-like | 766 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648853.1 | Tinospora cordifolia | succinate dehydrogenase subunit 7B, mitochondrial-like | 574 | F:GO:0043495; C:GO:0005749 | F:protein-membrane adaptor activity; C:mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | C:mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) | view details | |||
| JG648308.1 | Tinospora cordifolia | Ubiquitin-fold modifier-conjugating enzyme 1 | 703 | P:GO:1990592; F:GO:0061657 | P:protein K69-linked ufmylation; F:UFM1 conjugating enzyme activity | Transferases | P:protein ufmylation; F:UFM1 conjugating enzyme activity | view details | ||
| JG648815.1 | Tinospora cordifolia | CLAVATA3/ESR (CLE)-related protein 12 | 761 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| JG648297.1 | Tinospora cordifolia | 40S ribosomal protein S11 | 568 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650869.1 | Tinospora cordifolia | thaumatin-like protein | 773 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647784.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 806 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649813.1 | Tinospora cordifolia | zinc finger A20 and AN1 domain-containing stress-associated protein 6-like | 808 | F:GO:0003677; F:GO:0008270 | F:DNA binding; F:zinc ion binding | F:DNA binding; F:zinc ion binding | view details | |||
| JG646817.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 808 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650395.1 | Tinospora cordifolia | wound-induced protein 1 | 763 | no GO terms | view details | |||||
| JG648951.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 798 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646894.1 | Tinospora cordifolia | probable glucuronoxylan glucuronosyltransferase IRX7 | 704 | P:GO:0010417; F:GO:0016740; C:GO:0005794; C:GO:0016020 | P:glucuronoxylan biosynthetic process; F:transferase activity; C:Golgi apparatus; C:membrane | Transferases | P:protein glycosylation; F:glycosyltransferase activity | view details | ||
| JG650003.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 803 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645658.1 | Tinospora cordifolia | small nuclear ribonucleoprotein E | 512 | P:GO:0000387; F:GO:0003723; C:GO:0005682; C:GO:0005685; C:GO:0005686; C:GO:0005687; C:GO:0046540; C:GO:0071011 | P:spliceosomal snRNP assembly; F:RNA binding; C:U5 snRNP; C:U1 snRNP; C:U2 snRNP; C:U4 snRNP; C:U4/U6 x U5 tri-snRNP complex; C:precatalytic spliceosome | P:mRNA splicing, via spliceosome; C:spliceosomal complex | view details | |||
| JG647943.1 | Tinospora cordifolia | DSBA-like thioredoxin domain | 730 | F:GO:0016491 | F:oxidoreductase activity | Oxidoreductases | F:oxidoreductase activity | view details | ||
| JG650625.1 | Tinospora cordifolia | transaldolase isoform X2 | 796 | P:GO:0005975; P:GO:0006098; F:GO:0004801; C:GO:0005737 | P:carbohydrate metabolic process; P:pentose-phosphate shunt; F:transaldolase activity; C:cytoplasm | Transaldolase | P:carbohydrate metabolic process; P:pentose-phosphate shunt; F:transaldolase activity; C:cytoplasm | view details | ||
| JG646454.1 | Tinospora cordifolia | protein CREG1-like | 757 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649523.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 803 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649263.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 794 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646889.1 | Tinospora cordifolia | CD2 antigen cytoplasmic tail-binding protein 2-like | 681 | C:GO:0005682 | C:U5 snRNP | C:U5 snRNP | view details | |||
| JG646145.1 | Tinospora cordifolia | ubiquitin-40S ribosomal protein S27a | 713 | P:GO:0006412; P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0003735; F:GO:0031386; F:GO:0031625; F:GO:0046872; C:GO:0005634; C:GO:0005737; C:GO:0005840 | P:translation; P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:structural constituent of ribosome; F:protein tag; F:ubiquitin protein ligase binding; F:metal ion binding; C:nucleus; C:cytoplasm; C:ribosome | P:translation; F:structural constituent of ribosome; F:protein binding; C:ribosome | view details | |||
| JG646107.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 695 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646839.1 | Tinospora cordifolia | polyphenol oxidase | 671 | P:GO:0046148; F:GO:0004097; F:GO:0046872 | P:pigment biosynthetic process; F:catechol oxidase activity; F:metal ion binding | Catechol oxidase | no GO terms | view details | ||
| JG646537.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 557 | F:protein binding | view details | |||||
| JG650670.1 | Tinospora cordifolia | mitochondrial phosphate carrier protein 3, mitochondrial-like | 776 | P:GO:1990547; F:GO:0005315; C:GO:0031305 | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity; C:integral component of mitochondrial inner membrane | Translocases | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity | view details | ||
| JG645670.1 | Tinospora cordifolia | probable ADP-ribosylation factor GTPase-activating protein AGD6 | 652 | P:GO:0016192; P:GO:0050790; F:GO:0005096 | P:vesicle-mediated transport; P:regulation of catalytic activity; F:GTPase activator activity | P:vesicle-mediated transport; F:GTPase activator activity | view details | |||
| JG645964.1 | Tinospora cordifolia | patellin-3 | 814 | P:GO:0007049; P:GO:0051301; F:GO:0008289; C:GO:0005737; C:GO:0016020 | P:cell cycle; P:cell division; F:lipid binding; C:cytoplasm; C:membrane | F:lipid binding | view details | |||
| JG647781.1 | Tinospora cordifolia | 2-oxoglutarate-dependent dioxygenase DAO-like | 807 | P:GO:0009852; P:GO:0010252; P:GO:1901576; F:GO:0016705; F:GO:0046872; F:GO:0050302; F:GO:0051213 | P:auxin catabolic process; P:auxin homeostasis; P:organic substance biosynthetic process; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:metal ion binding; F:indole-3-acetaldehyde oxidase activity; F:dioxygenase activity | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2); Aryl-aldehyde oxidase; Indole-3-acetaldehyde oxidase; Aldehyde oxidase | no GO terms | view details | ||
| JG649149.1 | Tinospora cordifolia | cytochrome b5 | 777 | F:GO:0004128; F:GO:0009703; F:GO:0020037; F:GO:0046872; C:GO:0016021; C:GO:0043231 | F:cytochrome-b5 reductase activity, acting on NAD(P)H; F:nitrate reductase (NADH) activity; F:heme binding; F:metal ion binding; C:integral component of membrane; C:intracellular membrane-bounded organelle | Nitrate reductase (NAD(P)H); Nitrate reductase (NADH); Cytochrome-b5 reductase; Nitrate reductase | no GO terms | view details | ||
| JG647818.1 | Tinospora cordifolia | homeobox-leucine zipper protein ATHB-12-like | 803 | P:GO:0045893; F:GO:0043565; C:GO:0005634 | P:positive regulation of DNA-templated transcription; F:sequence-specific DNA binding; C:nucleus | P:regulation of DNA-templated transcription; F:DNA binding; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding | view details | |||
| JG650882.1 | Tinospora cordifolia | protein CREG1-like | 752 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649292.1 | Tinospora cordifolia | ferredoxin, root R-B2-like | 644 | P:GO:0022900; F:GO:0009055; F:GO:0046872; F:GO:0051537; C:GO:0009507 | P:electron transport chain; F:electron transfer activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; C:chloroplast | Oxidoreductases | P:electron transport chain; F:electron transfer activity; F:iron-sulfur cluster binding; F:2 iron, 2 sulfur cluster binding | view details | ||
| JG646998.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 757 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG646129.1 | Tinospora cordifolia | thaumatin-like protein | 783 | no GO terms | view details | |||||
| JG647777.1 | Tinospora cordifolia | zinc finger protein ZAT12-like | 805 | P:GO:0010200 | P:response to chitin | no GO terms | view details | |||
| JG648612.1 | Tinospora cordifolia | Sterile alpha motif domain-containing protein | 838 | F:GO:0005524; F:GO:0016491; F:GO:0051536 | F:ATP binding; F:oxidoreductase activity; F:iron-sulfur cluster binding | Oxidoreductases | F:protein binding | view details | ||
| JG645860.1 | Tinospora cordifolia | basic endochitinase-like | 867 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG650621.1 | Tinospora cordifolia | non-specific lipid transfer protein GPI-anchored 1-like | 782 | C:GO:0031224 | C:intrinsic component of membrane | no GO terms | view details | |||
| JG646446.1 | Tinospora cordifolia | thaumatin-like protein | 897 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648648.1 | Tinospora cordifolia | probable glutathione S-transferase | 811 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG649337.1 | Tinospora cordifolia | ---NA--- | 779 | no GO terms | view details | |||||
| JG646670.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 585 | F:protein binding | view details | |||||
| JG645662.1 | Tinospora cordifolia | O-glucosyltransferase rumi homolog | 781 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG649840.1 | Tinospora cordifolia | LRR receptor-like serine/threonine-protein kinase ERECTA | 786 | P:GO:0009987; F:GO:0005488; F:GO:0016740; C:GO:0016020 | P:cellular process; F:binding; F:transferase activity; C:membrane | Transferases | F:protein binding | view details | ||
| JG649548.1 | Tinospora cordifolia | glutathione S-transferase F9-like | 574 | P:GO:0006749; P:GO:0009407; F:GO:0004364; F:GO:0043295; C:GO:0005829 | P:glutathione metabolic process; P:toxin catabolic process; F:glutathione transferase activity; F:glutathione binding; C:cytosol | Glutathione transferase | no GO terms | view details | ||
| JG646602.1 | Tinospora cordifolia | small nuclear ribonucleoprotein SmD3b | 690 | P:GO:0000387; F:GO:0003723; C:GO:0000243; C:GO:0005682; C:GO:0005685; C:GO:0005686; C:GO:0005687; C:GO:0005689; C:GO:0005829; C:GO:0034715; C:GO:0034719; C:GO:0071011; C:GO:0071013; C:GO:0097526 | P:spliceosomal snRNP assembly; F:RNA binding; C:commitment complex; C:U5 snRNP; C:U1 snRNP; C:U2 snRNP; C:U4 snRNP; C:U12-type spliceosomal complex; C:cytosol; C:pICln-Sm protein complex; C:SMN-Sm protein complex; C:precatalytic spliceosome; C:catalytic step 2 spliceosome; C:spliceosomal tri-snRNP complex | P:spliceosomal snRNP assembly; P:RNA processing; C:spliceosomal complex | view details | |||
| JG648388.1 | Tinospora cordifolia | thaumatin-like protein | 798 | no GO terms | view details | |||||
| JG650936.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 592 | F:protein binding | view details | |||||
| JG649890.1 | Tinospora cordifolia | Transcription initiation factor tfiid subunit | 514 | C:GO:0110165 | C:cellular anatomical entity | no GO terms | view details | |||
| JG645886.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 805 | no GO terms | view details | |||||
| JG649387.1 | Tinospora cordifolia | Bet v I domain | 769 | F:GO:0016829; C:GO:0016020 | F:lyase activity; C:membrane | Lyases | P:defense response | view details | ||
| JG648879.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 648 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG647273.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 562 | F:protein binding | view details | |||||
| JG649838.1 | Tinospora cordifolia | derlin-2.2 | 854 | P:GO:0030433; P:GO:0030968; F:GO:0051787; F:GO:1990381; C:GO:0000839; C:GO:0030176 | P:ubiquitin-dependent ERAD pathway; P:endoplasmic reticulum unfolded protein response; F:misfolded protein binding; F:ubiquitin-specific protease binding; C:Hrd1p ubiquitin ligase ERAD-L complex; C:integral component of endoplasmic reticulum membrane | no GO terms | view details | |||
| JG649199.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 764 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647003.1 | Tinospora cordifolia | ubiquitin-60S ribosomal protein L40 | 647 | P:GO:0006412; P:GO:0016567; P:GO:0019941; F:GO:0003729; F:GO:0003735; F:GO:0031386; F:GO:0031625; C:GO:0005634; C:GO:0005840; C:GO:0009536 | P:translation; P:protein ubiquitination; P:modification-dependent protein catabolic process; F:mRNA binding; F:structural constituent of ribosome; F:protein tag; F:ubiquitin protein ligase binding; C:nucleus; C:ribosome; C:plastid | P:translation; F:structural constituent of ribosome; F:protein binding; C:ribosome | view details | |||
| JG648505.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 582 | F:protein binding | view details | |||||
| JG647233.1 | Tinospora cordifolia | 60S ribosomal protein L21-1 | 707 | P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648352.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 816 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648314.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 718 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645645.1 | Tinospora cordifolia | UPF0426 protein At1g28150, chloroplastic | 491 | F:GO:0003676 | F:nucleic acid binding | no GO terms | view details | |||
| JG647822.1 | Tinospora cordifolia | serine/threonine-protein kinase STY13 | 735 | P:GO:0006468; F:GO:0004674; F:GO:0005524 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG648660.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 746 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647296.1 | Tinospora cordifolia | 1-aminocyclopropane-1-carboxylate oxidase | 804 | P:GO:1901576; F:GO:0016705; F:GO:0046872; F:GO:0051213 | P:organic substance biosynthetic process; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:metal ion binding; F:dioxygenase activity | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | no GO terms | view details | ||
| JG647117.1 | Tinospora cordifolia | 40S ribosomal protein S10-1 | 790 | F:GO:0003723; F:GO:0003735; C:GO:0022627 | F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | no GO terms | view details | |||
| JG649215.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 890 | F:GO:0016881; C:GO:0005737 | F:acid-amino acid ligase activity; C:cytoplasm | Forming carbon-nitrogen bonds | no GO terms | view details | ||
| JG646226.1 | Tinospora cordifolia | Polygalacturonase inhibitor | 512 | no GO terms | view details | |||||
| JG649414.1 | Tinospora cordifolia | translationally-controlled tumor protein homolog | 803 | C:GO:0005737 | C:cytoplasm | no GO terms | view details | |||
| JG650755.1 | Tinospora cordifolia | sm-like protein LSM1B | 839 | P:GO:0000290; P:GO:0006397; F:GO:0000339; C:GO:0000932; C:GO:1990726 | P:deadenylation-dependent decapping of nuclear-transcribed mRNA; P:mRNA processing; F:RNA cap binding; C:P-body; C:Lsm1-7-Pat1 complex | P:nuclear-transcribed mRNA catabolic process | view details | |||
| JG647558.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 728 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645607.1 | Tinospora cordifolia | universal stress protein PHOS34 | 697 | F:GO:0016787 | F:hydrolase activity | Hydrolases | no GO terms | view details | ||
| JG646650.1 | Tinospora cordifolia | MOB kinase activator-like 1A | 792 | P:GO:0007165; P:GO:0032147; F:GO:0016301; F:GO:0030295; C:GO:0005634; C:GO:0005737 | P:signal transduction; P:activation of protein kinase activity; F:kinase activity; F:protein kinase activator activity; C:nucleus; C:cytoplasm | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG646499.1 | Tinospora cordifolia | ripening-related protein grip22 | 787 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647674.1 | Tinospora cordifolia | glucan endo-1,3-beta-glucosidase, basic isoform-like | 738 | P:GO:0005975; F:GO:0016765; F:GO:0042973; C:GO:0046658 | P:carbohydrate metabolic process; F:transferase activity, transferring alkyl or aryl (other than methyl) groups; F:glucan endo-1,3-beta-D-glucosidase activity; C:anchored component of plasma membrane | Transferring alkyl or aryl groups, other than methyl groups; Beta-glucosidase; Glucan endo-1,3-beta-D-glucosidase | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds | view details | ||
| JG647714.1 | Tinospora cordifolia | 60S ribosomal protein L18a-like | 701 | P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649158.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 805 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649237.1 | Tinospora cordifolia | lipid transfer-like protein vas | 712 | C:GO:0016020 | C:membrane | P:lipid transport; F:lipid binding | view details | |||
| JG648689.1 | Tinospora cordifolia | transcription factor bHLH106 | 741 | F:GO:0003677; F:GO:0046983; C:GO:0005634 | F:DNA binding; F:protein dimerization activity; C:nucleus | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity; F:protein dimerization activity | view details | |||
| JG647733.1 | Tinospora cordifolia | peroxisomal membrane protein 11C | 835 | P:GO:0016559; P:GO:0044375; F:GO:0042802; C:GO:0005779 | P:peroxisome fission; P:regulation of peroxisome size; F:identical protein binding; C:integral component of peroxisomal membrane | P:peroxisome fission; C:integral component of peroxisomal membrane | view details | |||
| JG647571.1 | Tinospora cordifolia | uncharacterized protein LOC122645445 isoform X1 | 793 | F:GO:0008092; C:GO:0005737 | F:cytoskeletal protein binding; C:cytoplasm | P:Arp2/3 complex-mediated actin nucleation; F:alpha-tubulin binding; C:early endosome; C:WASH complex | view details | |||
| JG648759.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 806 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647963.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 764 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646032.1 | Tinospora cordifolia | autophagy-related protein 8f | 726 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016020 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:membrane | no GO terms | view details | |||
| JG647053.1 | Tinospora cordifolia | putative dihydropyrimidinase | 748 | F:GO:0016810; C:GO:0005737 | F:hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds; C:cytoplasm | Acting on carbon-nitrogen bonds, other than peptide bonds | F:hydrolase activity; F:hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | view details | ||
| JG649412.1 | Tinospora cordifolia | transmembrane 9 superfamily member 8 | 834 | P:GO:0006817; P:GO:0050896; C:GO:0000139; C:GO:0005802; C:GO:0010008; C:GO:0016021 | P:phosphate ion transport; P:response to stimulus; C:Golgi membrane; C:trans-Golgi network; C:endosome membrane; C:integral component of membrane | C:integral component of membrane | view details | |||
| JG647100.1 | Tinospora cordifolia | 40S ribosomal protein S15 | 724 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022627 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG648902.1 | Tinospora cordifolia | ADP-ribosylation factor 2 | 800 | F:GO:0003924; F:GO:0005525 | F:GTPase activity; F:GTP binding | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG650665.1 | Tinospora cordifolia | N-(5-amino-5-carboxypentanoyl)-L-cysteinyl-D-valine synthase | 877 | F:GO:0003676 | F:nucleic acid binding | F:nucleic acid binding | view details | |||
| JG647107.1 | Tinospora cordifolia | Hypothetical predicted protein | 784 | P:GO:0006508; F:GO:0008233 | P:proteolysis; F:peptidase activity | Acting on peptide bonds (peptidases) | no GO terms | view details | ||
| JG649107.1 | Tinospora cordifolia | 60S ribosomal protein L18a | 681 | P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG645686.1 | Tinospora cordifolia | 40S ribosomal protein S18 | 703 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0005829; C:GO:0015935 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosol; C:small ribosomal subunit | P:translation; F:nucleic acid binding; F:RNA binding; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648246.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 623 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649538.1 | Tinospora cordifolia | outer envelope pore protein 16-3, chloroplastic/mitochondrial | 676 | P:GO:0045039; C:GO:0016021; C:GO:0042721 | P:protein insertion into mitochondrial inner membrane; C:integral component of membrane; C:TIM22 mitochondrial import inner membrane insertion complex | P:protein insertion into mitochondrial inner membrane; C:TIM22 mitochondrial import inner membrane insertion complex | view details | |||
| JG649701.1 | Tinospora cordifolia | 60S ribosomal protein L22-2-like | 569 | P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0005737; C:GO:0005840 | P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytoplasm; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650709.1 | Tinospora cordifolia | anaphase-promoting complex subunit 1 isoform X2 | 863 | C:GO:0005680 | C:anaphase-promoting complex | C:anaphase-promoting complex | view details | |||
| JG645566.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 809 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647992.1 | Tinospora cordifolia | 40S ribosomal protein S7 | 779 | P:GO:0006364; P:GO:0006412; P:GO:0042274; F:GO:0003735; C:GO:0022627; C:GO:0032040 | P:rRNA processing; P:translation; P:ribosomal small subunit biogenesis; F:structural constituent of ribosome; C:cytosolic small ribosomal subunit; C:small-subunit processome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649545.1 | Tinospora cordifolia | transcription factor MYB108-like | 625 | P:GO:0006355; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | no GO terms | view details | |||
| JG650581.1 | Tinospora cordifolia | thaumatin-like protein | 841 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG647799.1 | Tinospora cordifolia | LOB domain-containing protein 15 | 813 | no GO terms | view details | |||||
| JG648453.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 612 | F:protein binding | view details | |||||
| JG647307.1 | Tinospora cordifolia | 60S ribosomal protein L14-1-like | 570 | P:GO:0006412; P:GO:0042273; F:GO:0003723; F:GO:0003735; C:GO:0022625; C:GO:0043231 | P:translation; P:ribosomal large subunit biogenesis; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit; C:intracellular membrane-bounded organelle | P:translation; F:RNA binding; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG649288.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 589 | F:protein binding | view details | |||||
| JG648050.1 | Tinospora cordifolia | major strawberry allergen Fra a 1-2-like | 730 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650891.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 655 | F:protein binding | view details | |||||
| JG647900.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 612 | F:protein binding | view details | |||||
| JG650055.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 688 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648302.1 | Tinospora cordifolia | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic | 712 | P:GO:0008299; P:GO:0019288; F:GO:0003824; F:GO:0016740; F:GO:0016779; F:GO:0050518; F:GO:0070567 | P:isoprenoid biosynthetic process; P:isopentenyl diphosphate biosynthetic process, methylerythritol 4-phosphate pathway; F:catalytic activity; F:transferase activity; F:nucleotidyltransferase activity; F:2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase activity; F:cytidylyltransferase activity | no GO terms | view details | |||
| JG645631.1 | Tinospora cordifolia | thaumatin-like protein | 845 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648148.1 | Tinospora cordifolia | 60S ribosomal protein L27 | 679 | P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647837.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 523 | F:protein binding | view details | |||||
| JG648114.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 603 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG649769.1 | Tinospora cordifolia | catechol O-methyltransferase | 756 | P:GO:0019438; P:GO:0032259; F:GO:0016206; F:GO:0046983; F:GO:0102084; F:GO:0102938 | P:aromatic compound biosynthetic process; P:methylation; F:catechol O-methyltransferase activity; F:protein dimerization activity; F:L-dopa O-methyltransferase activity; F:orcinol O-methyltransferase activity | Catechol O-methyltransferase | F:methyltransferase activity; F:O-methyltransferase activity | view details | ||
| JG646518.1 | Tinospora cordifolia | thaumatin-like protein | 826 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG646533.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 711 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648179.1 | Tinospora cordifolia | thaumatin-like protein | 804 | no GO terms | view details | |||||
| JG646996.1 | Tinospora cordifolia | CASP-like protein 1D1 | 635 | F:GO:0051539; C:GO:0005886 | F:4 iron, 4 sulfur cluster binding; C:plasma membrane | no GO terms | view details | |||
| JG645560.1 | Tinospora cordifolia | retrovirus-related Pol polyprotein from transposon TNT 1-94 | 870 | P:GO:0050896; P:GO:0055085; F:GO:0022857; F:GO:0043167; F:GO:0097159; F:GO:1901363; C:GO:0016020 | P:response to stimulus; P:transmembrane transport; F:transmembrane transporter activity; F:ion binding; F:organic cyclic compound binding; F:heterocyclic compound binding; C:membrane | Translocases | no GO terms | view details | ||
| JG648529.1 | Tinospora cordifolia | thaumatin-like protein | 817 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646077.1 | Tinospora cordifolia | Glyoxalase-like domain | 798 | no GO terms | view details | |||||
| JG648674.1 | Tinospora cordifolia | nuclear transport factor 2B | 801 | P:GO:0006913; C:GO:0044613 | P:nucleocytoplasmic transport; C:nuclear pore central transport channel | P:nucleocytoplasmic transport | view details | |||
| JG648417.1 | Tinospora cordifolia | 40S ribosomal protein S23 | 706 | P:GO:0006412; F:GO:0003735; C:GO:0016021; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:integral component of membrane; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG647557.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 792 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650088.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 664 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650551.1 | Tinospora cordifolia | 60S ribosomal protein L35 | 792 | P:GO:0000463; P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0022625 | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA); P:translation; F:structural constituent of ribosome; C:ribosome; C:cytosolic large ribosomal subunit | view details | |||
| JG646714.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 810 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649773.1 | Tinospora cordifolia | probable indole-3-acetic acid-amido synthetase GH3.1 | 752 | no GO terms | view details | |||||
| JG648483.1 | Tinospora cordifolia | basic endochitinase-like | 880 | P:GO:0005975; P:GO:0006032; P:GO:0016998; P:GO:0050832; F:GO:0004568; F:GO:0008061 | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; P:defense response to fungus; F:chitinase activity; F:chitin binding | Chitinase | P:carbohydrate metabolic process; P:chitin catabolic process; P:cell wall macromolecule catabolic process; F:chitinase activity | view details | ||
| JG646640.1 | Tinospora cordifolia | probable WRKY transcription factor 75 | 821 | P:GO:0006355; F:GO:0003700; F:GO:0043565; C:GO:0005634 | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding; C:nucleus | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity; F:sequence-specific DNA binding | view details | |||
| JG647679.1 | Tinospora cordifolia | 60S ribosomal protein L34 | 609 | P:GO:0006412; P:GO:0042254; F:GO:0003735; C:GO:0022625 | P:translation; P:ribosome biogenesis; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647866.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 805 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650930.1 | Tinospora cordifolia | cysteine proteinase inhibitor 12 | 580 | P:GO:0010951; F:GO:0004869; C:GO:0016021 | P:negative regulation of endopeptidase activity; F:cysteine-type endopeptidase inhibitor activity; C:integral component of membrane | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG650053.1 | Tinospora cordifolia | putative disease resistance protein RGA4 | 590 | P:GO:0006952; P:GO:0006955; P:GO:0098542; F:GO:0043531 | P:defense response; P:immune response; P:defense response to other organism; F:ADP binding | no GO terms | view details | |||
| JG649678.1 | Tinospora cordifolia | protein CREG1-like | 756 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650362.1 | Tinospora cordifolia | RWD domain-containing protein 1 | 848 | P:GO:0002181; C:GO:0005844 | P:cytoplasmic translation; C:polysome | F:protein binding | view details | |||
| JG649063.1 | Tinospora cordifolia | histidinol dehydrogenase, chloroplastic isoform X1 | 800 | P:GO:0000105; P:GO:0009411; P:GO:0009555; F:GO:0004399; F:GO:0046872; F:GO:0051287; C:GO:0005829; C:GO:0009507 | P:histidine biosynthetic process; P:response to UV; P:pollen development; F:histidinol dehydrogenase activity; F:metal ion binding; F:NAD binding; C:cytosol; C:chloroplast | Histidinol dehydrogenase | F:oxidoreductase activity; F:oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; F:metal ion binding; F:NAD binding | view details | ||
| JG650706.1 | Tinospora cordifolia | eukaryotic translation initiation factor 5A | 792 | P:GO:0006413; P:GO:0045901; P:GO:0045905; F:GO:0003723; F:GO:0003743; F:GO:0003746; F:GO:0043022 | P:translational initiation; P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation initiation factor activity; F:translation elongation factor activity; F:ribosome binding | P:positive regulation of translational elongation; P:positive regulation of translational termination; F:RNA binding; F:translation elongation factor activity; F:ribosome binding | view details | |||
| JG648262.1 | Tinospora cordifolia | ripening-related protein grip22 | 765 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG645830.1 | Tinospora cordifolia | cytochrome c oxidase subunit 6b-1-like | 851 | C:GO:0005739; C:GO:0045277 | C:mitochondrion; C:respiratory chain complex IV | C:mitochondrion; C:respiratory chain complex IV | view details | |||
| JG647381.1 | Tinospora cordifolia | cytochrome P450 78A7-like | 618 | P:GO:0033075; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:isoquinoline alkaloid biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645567.1 | Tinospora cordifolia | translationally-controlled tumor protein homolog | 731 | C:GO:0005737 | C:cytoplasm | no GO terms | view details | |||
| JG645865.1 | Tinospora cordifolia | Retrovirus-related Pol polyprotein from transposon RE1 | 822 | P:GO:0015074; F:GO:0003676; F:GO:0008270 | P:DNA integration; F:nucleic acid binding; F:zinc ion binding | no GO terms | view details | |||
| JG646630.1 | Tinospora cordifolia | siRNA-mediated silencing protein NRDE-2 | 844 | C:GO:0000178; C:GO:0005634 | C:exosome (RNase complex); C:nucleus | no GO terms | view details | |||
| JG645613.1 | Tinospora cordifolia | probable chalcone--flavonone isomerase 3 | 768 | P:GO:0009813; F:GO:0016872 | P:flavonoid biosynthetic process; F:intramolecular lyase activity | Intramolecular lyases | F:intramolecular lyase activity | view details | ||
| JG649855.1 | Tinospora cordifolia | protein transport protein Sec23A-like | 768 | P:GO:0006886; P:GO:0050790; P:GO:0090110; F:GO:0005096; F:GO:0008270; C:GO:0005789; C:GO:0030127; C:GO:0070971 | P:intracellular protein transport; P:regulation of catalytic activity; P:COPII-coated vesicle cargo loading; F:GTPase activator activity; F:zinc ion binding; C:endoplasmic reticulum membrane; C:COPII vesicle coat; C:endoplasmic reticulum exit site | P:intracellular protein transport; P:endoplasmic reticulum to Golgi vesicle-mediated transport; P:COPII-coated vesicle budding; C:COPII vesicle coat | view details | |||
| JG646059.1 | Tinospora cordifolia | 40S ribosomal protein S23 | 750 | P:GO:0006412; F:GO:0003735; C:GO:0016021; C:GO:0022627 | P:translation; F:structural constituent of ribosome; C:integral component of membrane; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome; C:small ribosomal subunit | view details | |||
| JG647782.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 803 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648198.1 | Tinospora cordifolia | thaumatin-like protein | 794 | no GO terms | view details | |||||
| JG650380.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 585 | F:protein binding | view details | |||||
| JG646620.1 | Tinospora cordifolia | integrin-linked protein kinase 1 | 649 | P:GO:0006468; F:GO:0004674; F:GO:0005524 | P:protein phosphorylation; F:protein serine/threonine kinase activity; F:ATP binding | Transferring phosphorus-containing groups | P:protein phosphorylation; F:protein kinase activity; F:ATP binding | view details | ||
| JG650752.1 | Tinospora cordifolia | DNA-directed RNA polymerase III subunit RPC6-like | 855 | F:GO:0016740; C:GO:0005666 | F:transferase activity; C:RNA polymerase III complex | Transferases | P:transcription by RNA polymerase III; C:RNA polymerase III complex | view details | ||
| JG648823.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 566 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650376.1 | Tinospora cordifolia | thaumatin-like protein | 818 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG650898.1 | Tinospora cordifolia | copper transport protein ATX1-like | 534 | F:GO:0046872 | F:metal ion binding | F:metal ion binding | view details | |||
| JG650146.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 691 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646684.1 | Tinospora cordifolia | ribonuclease 3 | 850 | P:GO:0006401; P:GO:0090502; F:GO:0003723; F:GO:0033897; C:GO:0005576 | P:RNA catabolic process; P:RNA phosphodiester bond hydrolysis, endonucleolytic; F:RNA binding; F:ribonuclease T2 activity; C:extracellular region | Acting on ester bonds; Ribonuclease T(2) | F:RNA binding; F:ribonuclease T2 activity | view details | ||
| JG645757.1 | Tinospora cordifolia | thaumatin-like protein | 824 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646726.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 799 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646587.1 | Tinospora cordifolia | thaumatin-like protein | 799 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647667.1 | Tinospora cordifolia | PITH domain-containing protein At3g04780 | 760 | no GO terms | view details | |||||
| JG645571.1 | Tinospora cordifolia | Glutathione S-transferase DHAR2 | 730 | P:GO:0010731; P:GO:0033355; P:GO:0098869; F:GO:0004364; F:GO:0045174 | P:protein glutathionylation; P:ascorbate glutathione cycle; P:cellular oxidant detoxification; F:glutathione transferase activity; F:glutathione dehydrogenase (ascorbate) activity | Glutathione transferase; Glutathione dehydrogenase (ascorbate) | P:ascorbate glutathione cycle; P:cellular oxidant detoxification; F:glutathione dehydrogenase (ascorbate) activity | view details | ||
| JG646540.1 | Tinospora cordifolia | protein RALF-like 33 | 659 | P:GO:0019722; C:GO:0005576 | P:calcium-mediated signaling; C:extracellular region | no GO terms | view details | |||
| JG650844.1 | Tinospora cordifolia | thaumatin-like protein | 770 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650684.1 | Tinospora cordifolia | thaumatin-like protein | 820 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG648564.1 | Tinospora cordifolia | Glycoside hydrolase | 795 | P:GO:0005975; F:GO:0004553; F:GO:0030246 | P:carbohydrate metabolic process; F:hydrolase activity, hydrolyzing O-glycosyl compounds; F:carbohydrate binding | Glycosylases | no GO terms | view details | ||
| JG649707.1 | Tinospora cordifolia | ABC transporter G family member 28-like isoform X2 | 756 | P:GO:0055085; F:GO:0005524; F:GO:0140359; C:GO:0016021 | P:transmembrane transport; F:ATP binding; F:ABC-type transporter activity; C:integral component of membrane | Translocases | F:ABC-type transporter activity | view details | ||
| JG649668.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 778 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG650049.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2-17 kDa | 723 | P:GO:0000209; P:GO:0006511; F:GO:0004839; F:GO:0005524; F:GO:0016746; F:GO:0061631; C:GO:0005634 | P:protein polyubiquitination; P:ubiquitin-dependent protein catabolic process; F:ubiquitin activating enzyme activity; F:ATP binding; F:acyltransferase activity; F:ubiquitin conjugating enzyme activity; C:nucleus | E1 ubiquitin-activating enzyme; E2 ubiquitin-conjugating enzyme | no GO terms | view details | ||
| JG647180.1 | Tinospora cordifolia | autophagy-related protein 8C-like | 706 | P:GO:0006914; P:GO:0006995; C:GO:0005874; C:GO:0016021 | P:autophagy; P:cellular response to nitrogen starvation; C:microtubule; C:integral component of membrane | no GO terms | view details | |||
| JG648310.1 | Tinospora cordifolia | transcription factor PIF3 isoform X1 | 815 | P:GO:0006355; F:GO:0003700; F:GO:0046983; C:GO:0005634; C:GO:0016020; C:GO:0016021 | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity; F:protein dimerization activity; C:nucleus; C:membrane; C:integral component of membrane | P:regulation of DNA-templated transcription; F:DNA-binding transcription factor activity | view details | |||
| JG648639.1 | Tinospora cordifolia | ankyrin repeat-containing protein NPR4-like | 668 | C:GO:0016020 | C:membrane | F:protein binding | view details | |||
| JG650308.1 | Tinospora cordifolia | protein CREG1-like | 846 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG647026.1 | Tinospora cordifolia | tetratricopeptide repeat protein 1-like | 786 | F:protein binding | view details | |||||
| JG650017.1 | Tinospora cordifolia | probable calcium-binding protein CML36 | 847 | F:GO:0005509 | F:calcium ion binding | F:calcium ion binding | view details | |||
| JG648747.1 | Tinospora cordifolia | protein CREG1 | 678 | C:GO:0005576; C:GO:0016020 | C:extracellular region; C:membrane | no GO terms | view details | |||
| JG645980.1 | Tinospora cordifolia | U1 small nuclear ribonucleoprotein 70 kDa | 855 | P:GO:0000398; F:GO:0003729; F:GO:0030619; C:GO:0005685; C:GO:0071004 | P:mRNA splicing, via spliceosome; F:mRNA binding; F:U1 snRNA binding; C:U1 snRNP; C:U2-type prespliceosome | F:nucleic acid binding; F:RNA binding | view details | |||
| JG648764.1 | Tinospora cordifolia | ---NA--- | 765 | no GO terms | view details | |||||
| JG647901.1 | Tinospora cordifolia | PLAT domain-containing protein 2-like | 592 | F:protein binding | view details | |||||
| JG647566.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 802 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648333.1 | Tinospora cordifolia | pathogen-related protein-like | 726 | no GO terms | view details | |||||
| JG645914.1 | Tinospora cordifolia | Udp-glycosyltransferase 87a2 | 707 | F:GO:0008194 | F:UDP-glycosyltransferase activity | Glycosyltransferases | F:UDP-glycosyltransferase activity | view details | ||
| JG647982.1 | Tinospora cordifolia | Fumarate hydratase 1, mitochondrial | 818 | P:GO:0006099; P:GO:0006106; P:GO:0006108; F:GO:0004333; C:GO:0005739; C:GO:0045239 | P:tricarboxylic acid cycle; P:fumarate metabolic process; P:malate metabolic process; F:fumarate hydratase activity; C:mitochondrion; C:tricarboxylic acid cycle enzyme complex | Fumarate hydratase | P:tricarboxylic acid cycle; P:fumarate metabolic process; F:catalytic activity; F:fumarate hydratase activity; F:lyase activity; C:tricarboxylic acid cycle enzyme complex | view details | ||
| JG650968.1 | Tinospora cordifolia | nucleoside diphosphate kinase B | 685 | P:GO:0006165; P:GO:0006183; P:GO:0006228; P:GO:0006241; F:GO:0004550; F:GO:0005524 | P:nucleoside diphosphate phosphorylation; P:GTP biosynthetic process; P:UTP biosynthetic process; P:CTP biosynthetic process; F:nucleoside diphosphate kinase activity; F:ATP binding | Nucleoside-diphosphate kinase | P:nucleoside diphosphate phosphorylation; P:GTP biosynthetic process; P:UTP biosynthetic process; P:CTP biosynthetic process; F:nucleoside diphosphate kinase activity | view details | ||
| JG650929.1 | Tinospora cordifolia | 26S protease regulatory subunit 7 | 759 | P:GO:0043161; P:GO:1901800; F:GO:0005524; F:GO:0008233; F:GO:0016887; F:GO:0036402; C:GO:0005634; C:GO:0005737; C:GO:0008540 | P:proteasome-mediated ubiquitin-dependent protein catabolic process; P:positive regulation of proteasomal protein catabolic process; F:ATP binding; F:peptidase activity; F:ATP hydrolysis activity; F:proteasome-activating activity; C:nucleus; C:cytoplasm; C:proteasome regulatory particle, base subcomplex | Acting on peptide bonds (peptidases); Proteasome ATPase; Nucleoside-triphosphate phosphatase | F:ATP binding; F:ATP hydrolysis activity | view details | ||
| JG647593.1 | Tinospora cordifolia | ripening-related protein grip22 | 758 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG650980.1 | Tinospora cordifolia | ---NA--- | 587 | no GO terms | view details | |||||
| JG650791.1 | Tinospora cordifolia | thaumatin-like protein | 830 | no GO terms | view details | |||||
| JG650839.1 | Tinospora cordifolia | protein RER1B-like | 843 | P:GO:0006621; P:GO:0006890; C:GO:0005783; C:GO:0030173 | P:protein retention in ER lumen; P:retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum; C:endoplasmic reticulum; C:integral component of Golgi membrane | C:integral component of membrane | view details | |||
| JG649424.1 | Tinospora cordifolia | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | 863 | P:GO:0032259; F:GO:0030776; F:GO:0030794; C:GO:0005737 | P:methylation; F:(RS)-1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase activity; F:(S)-coclaurine-N-methyltransferase activity; C:cytoplasm | (S)-coclaurine-N-methyltransferase; (RS)-1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase | no GO terms | view details | ||
| JG646181.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 573 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG649860.1 | Tinospora cordifolia | thaumatin-like protein | 792 | no GO terms | view details | |||||
| JG648487.1 | Tinospora cordifolia | thaumatin-like protein | 893 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649236.1 | Tinospora cordifolia | ripening-related protein grip22 | 741 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649688.1 | Tinospora cordifolia | Pyrrolidone-carboxylate peptidase | 846 | P:GO:0006508; F:GO:0016920; C:GO:0005829 | P:proteolysis; F:pyroglutamyl-peptidase activity; C:cytosol | Acting on peptide bonds (peptidases) | P:proteolysis; F:pyroglutamyl-peptidase activity; C:cytosol | view details | ||
| JG645666.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 801 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647501.1 | Tinospora cordifolia | probable glutathione S-transferase | 770 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG649844.1 | Tinospora cordifolia | ripening-related protein grip22 | 773 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646463.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 808 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG649997.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 773 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648947.1 | Tinospora cordifolia | Glutamyl-tRNA(Gln) amidotransferase subunit D like | 678 | F:GO:0016740; C:GO:0016021 | F:transferase activity; C:integral component of membrane | Transferases | no GO terms | view details | ||
| JG649168.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 789 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG645994.1 | Tinospora cordifolia | histone H2B-like | 765 | P:GO:0006334; F:GO:0003677; F:GO:0046982; C:GO:0000786; C:GO:0005634 | P:nucleosome assembly; F:DNA binding; F:protein heterodimerization activity; C:nucleosome; C:nucleus | F:DNA binding; F:structural constituent of chromatin; F:protein heterodimerization activity; C:nucleosome | view details | |||
| JG646565.1 | Tinospora cordifolia | serine/threonine-protein kinase Nek6-like | 743 | P:GO:0016310; F:GO:0000166; F:GO:0004672 | P:phosphorylation; F:nucleotide binding; F:protein kinase activity | Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG650496.1 | Tinospora cordifolia | heat shock factor-binding protein 1-like | 673 | P:GO:0045892; P:GO:0048316; P:GO:0070370; F:GO:0003714; F:GO:0043621; C:GO:0005634; C:GO:0005829 | P:negative regulation of DNA-templated transcription; P:seed development; P:cellular heat acclimation; F:transcription corepressor activity; F:protein self-association; C:nucleus; C:cytosol | F:transcription corepressor activity | view details | |||
| JG649503.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 808 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649564.1 | Tinospora cordifolia | cytochrome P450 CYP82D47-like | 841 | P:GO:0019438; P:GO:0098542; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037; C:GO:0016021 | P:aromatic compound biosynthetic process; P:defense response to other organism; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding; C:integral component of membrane | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650186.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 674 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646766.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 803 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650699.1 | Tinospora cordifolia | protein CREG1-like | 674 | C:GO:0005576; C:GO:0016020 | C:extracellular region; C:membrane | no GO terms | view details | |||
| JG646283.1 | Tinospora cordifolia | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ | 610 | P:GO:0006633; P:GO:0009245; F:GO:0004317; F:GO:0008659; F:GO:0008693; F:GO:0047451; C:GO:0005737 | P:fatty acid biosynthetic process; P:lipid A biosynthetic process; F:3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase activity; F:(3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase activity; F:3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase activity; F:3-hydroxyoctanoyl-[acyl-carrier-protein] dehydratase activity; C:cytoplasm | Fatty-acid synthase system; 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | no GO terms | view details | ||
| JG646751.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 802 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG646615.1 | Tinospora cordifolia | RNA/RNP complex-1-interacting phosphatase | 710 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650198.1 | Tinospora cordifolia | nudix hydrolase 2 | 712 | F:GO:0035529; F:GO:0047631; F:GO:0051287 | F:NADH pyrophosphatase activity; F:ADP-ribose diphosphatase activity; F:NAD binding | NAD(+) diphosphatase; ADP-ribose diphosphatase | no GO terms | view details | ||
| JG650700.1 | Tinospora cordifolia | Mediator of RNA polymerase II transcription subunit like | 707 | no GO terms | view details | |||||
| JG647729.1 | Tinospora cordifolia | cell number regulator 8-like | 828 | no GO terms | view details | |||||
| JG647009.1 | Tinospora cordifolia | RING-H2 finger protein ATL66-like | 913 | P:GO:0016567; C:GO:0016020; C:GO:0016021 | P:protein ubiquitination; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG649999.1 | Tinospora cordifolia | GTP-binding nuclear protein Ran-3 | 728 | P:GO:0000054; P:GO:0006606; F:GO:0003924; F:GO:0005525; F:GO:0016829; C:GO:0005634; C:GO:0005737 | P:ribosomal subunit export from nucleus; P:protein import into nucleus; F:GTPase activity; F:GTP binding; F:lyase activity; C:nucleus; C:cytoplasm | Lyases; Nucleoside-triphosphate phosphatase | P:nucleocytoplasmic transport; F:GTPase activity; F:GTP binding | view details | ||
| JG645796.1 | Tinospora cordifolia | protein CREG1 | 755 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648029.1 | Tinospora cordifolia | 40S ribosomal protein S5 | 788 | P:GO:0000028; P:GO:0006412; F:GO:0003729; F:GO:0003735; F:GO:0019843; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:mRNA binding; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic small ribosomal subunit | P:translation; F:structural constituent of ribosome; C:small ribosomal subunit | view details | |||
| JG647889.1 | Tinospora cordifolia | 60S ribosomal protein L36-2-like | 759 | P:GO:0006412; F:GO:0003735; C:GO:0005840 | P:translation; F:structural constituent of ribosome; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650029.1 | Tinospora cordifolia | hypothetical protein BVC80_665g4 | 760 | C:GO:0016020; C:GO:0016021 | C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG647506.1 | Tinospora cordifolia | NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial | 712 | P:GO:0009060; P:GO:0015990; P:GO:0022900; P:GO:0032981; F:GO:0008137; F:GO:0046872; F:GO:0048038; F:GO:0051539; C:GO:0005747 | P:aerobic respiration; P:electron transport coupled proton transport; P:electron transport chain; P:mitochondrial respiratory chain complex I assembly; F:NADH dehydrogenase (ubiquinone) activity; F:metal ion binding; F:quinone binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrial respiratory chain complex I | NADH:ubiquinone reductase (H(+)-translocating); NADH dehydrogenase (quinone); NAD(P)H dehydrogenase (quinone) | F:NADH dehydrogenase (ubiquinone) activity; F:quinone binding; F:iron-sulfur cluster binding; F:4 iron, 4 sulfur cluster binding | view details | ||
| JG648494.1 | Tinospora cordifolia | B2 protein | 700 | P:GO:0034976 | P:response to endoplasmic reticulum stress | P:response to endoplasmic reticulum stress | view details | |||
| JG647250.1 | Tinospora cordifolia | arogenate dehydrogenase 2, chloroplastic-like | 820 | P:GO:0006571; F:GO:0004665; F:GO:0008977; F:GO:0033730 | P:tyrosine biosynthetic process; F:prephenate dehydrogenase (NADP+) activity; F:prephenate dehydrogenase (NAD+) activity; F:arogenate dehydrogenase (NADP+) activity | Arogenate dehydrogenase (NADP(+)); Prephenate dehydrogenase; Prephenate dehydrogenase (NADP(+)); Arogenate dehydrogenase (NAD(P)(+)) | P:tyrosine biosynthetic process; F:prephenate dehydrogenase (NADP+) activity; F:prephenate dehydrogenase (NAD+) activity; F:arogenate dehydrogenase (NADP+) activity | view details | ||
| JG647069.1 | Tinospora cordifolia | pathogen-related protein-like | 845 | no GO terms | view details | |||||
| JG649112.1 | Tinospora cordifolia | thaumatin-like protein | 819 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG648681.1 | Tinospora cordifolia | PLAT domain-containing protein 1-like | 585 | F:protein binding | view details | |||||
| JG650515.1 | Tinospora cordifolia | ripening-related protein grip22 | 818 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG646547.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 588 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG646041.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 810 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG646105.1 | Tinospora cordifolia | Fiber protein Fb34 | 724 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG645765.1 | Tinospora cordifolia | nudix hydrolase 16, mitochondrial | 715 | F:GO:0016787; C:GO:0005634; C:GO:0005737 | F:hydrolase activity; C:nucleus; C:cytoplasm | Hydrolases | no GO terms | view details | ||
| JG649676.1 | Tinospora cordifolia | succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial-like | 785 | P:GO:0006099; P:GO:0006124; P:GO:0022904; F:GO:0008177; F:GO:0009055; F:GO:0046872; F:GO:0051537; F:GO:0051538; F:GO:0051539; C:GO:0005743; C:GO:0045273 | P:tricarboxylic acid cycle; P:ferredoxin metabolic process; P:respiratory electron transport chain; F:succinate dehydrogenase (ubiquinone) activity; F:electron transfer activity; F:metal ion binding; F:2 iron, 2 sulfur cluster binding; F:3 iron, 4 sulfur cluster binding; F:4 iron, 4 sulfur cluster binding; C:mitochondrial inner membrane; C:respiratory chain complex II | Succinate dehydrogenase (quinone) | P:tricarboxylic acid cycle; F:electron transfer activity; F:oxidoreductase activity; F:iron-sulfur cluster binding | view details | ||
| JG647521.1 | Tinospora cordifolia | mitochondrial import receptor subunit TOM20-like | 738 | P:GO:0045040; F:GO:0046872; C:GO:0005742; C:GO:0016021 | P:protein insertion into mitochondrial outer membrane; F:metal ion binding; C:mitochondrial outer membrane translocase complex; C:integral component of membrane | P:protein insertion into mitochondrial outer membrane; F:protein binding; C:mitochondrial outer membrane translocase complex | view details | |||
| JG647683.1 | Tinospora cordifolia | probable 2-oxoglutarate-dependent dioxygenase At5g05600 | 761 | F:GO:0046872; F:GO:0051213 | F:metal ion binding; F:dioxygenase activity | Oxidoreductases | no GO terms | view details | ||
| JG646189.1 | Tinospora cordifolia | hypothetical protein IFM89_003051 | 505 | no GO terms | view details | |||||
| JG650086.1 | Tinospora cordifolia | 60S ribosomal protein L23A | 625 | P:GO:0000027; P:GO:0006412; F:GO:0003729; F:GO:0003735; F:GO:0019843; C:GO:0022625 | P:ribosomal large subunit assembly; P:translation; F:mRNA binding; F:structural constituent of ribosome; F:rRNA binding; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647458.1 | Tinospora cordifolia | SUMO-conjugating enzyme SCE1 | 697 | P:GO:0016925; F:GO:0005524; F:GO:0061656; C:GO:0005634 | P:protein sumoylation; F:ATP binding; F:SUMO conjugating enzyme activity; C:nucleus | Transferases | no GO terms | view details | ||
| JG646825.1 | Tinospora cordifolia | leucine-rich repeat protein 1-like | 763 | P:GO:0016310; F:GO:0016301; F:GO:0032440 | P:phosphorylation; F:kinase activity; F:2-alkenal reductase [NAD(P)+] activity | 2-alkenal reductase (NAD(P)(+)); Transferring phosphorus-containing groups | F:protein binding | view details | ||
| JG649756.1 | Tinospora cordifolia | protein RALF-like 33 | 701 | P:GO:0019722; C:GO:0005576 | P:calcium-mediated signaling; C:extracellular region | no GO terms | view details | |||
| JG649487.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 649 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG646396.1 | Tinospora cordifolia | protein TIFY 3B | 846 | P:GO:0009611; P:GO:0031347; P:GO:2000022; C:GO:0005634 | P:response to wounding; P:regulation of defense response; P:regulation of jasmonic acid mediated signaling pathway; C:nucleus | no GO terms | view details | |||
| JG648100.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 712 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650717.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 810 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG647635.1 | Tinospora cordifolia | LOC110425786 isoform | 862 | no GO terms | view details | |||||
| JG650383.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 815 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648033.1 | Tinospora cordifolia | probable glutathione S-transferase | 848 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | P:glutathione metabolic process; F:glutathione transferase activity | view details | ||
| JG648021.1 | Tinospora cordifolia | Mannose-binding lectin | 639 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG645576.1 | Tinospora cordifolia | actin-depolymerizing factor 7 | 763 | P:GO:0030042; F:GO:0051015; C:GO:0005737; C:GO:0015629 | P:actin filament depolymerization; F:actin filament binding; C:cytoplasm; C:actin cytoskeleton | P:actin filament depolymerization; F:actin binding; C:actin cytoskeleton | view details | |||
| JG650990.1 | Tinospora cordifolia | thaumatin-like protein | 829 | no GO terms | view details | |||||
| JG650794.1 | Tinospora cordifolia | subtilisin-like protease SBT3.17 | 539 | P:GO:0006508; P:GO:0016310; F:GO:0008233; F:GO:0016301 | P:proteolysis; P:phosphorylation; F:peptidase activity; F:kinase activity | Acting on peptide bonds (peptidases); Transferring phosphorus-containing groups | no GO terms | view details | ||
| JG647160.1 | Tinospora cordifolia | 60S ribosomal protein L6 | 825 | P:GO:0000027; P:GO:0002181; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:ribosomal large subunit assembly; P:cytoplasmic translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG647964.1 | Tinospora cordifolia | probable chalcone--flavonone isomerase 3 | 709 | P:GO:0009813; F:GO:0016872 | P:flavonoid biosynthetic process; F:intramolecular lyase activity | Intramolecular lyases | F:intramolecular lyase activity | view details | ||
| JG646240.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 684 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG650901.1 | Tinospora cordifolia | copper transport protein ATX1-like | 506 | F:GO:0046872 | F:metal ion binding | F:metal ion binding | view details | |||
| JG649224.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 784 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG649781.1 | Tinospora cordifolia | inosine triphosphate pyrophosphatase | 697 | P:GO:0009117; P:GO:0009204; P:GO:0034404; F:GO:0000166; F:GO:0035529; F:GO:0036218; F:GO:0046872; C:GO:0005737 | P:nucleotide metabolic process; P:deoxyribonucleoside triphosphate catabolic process; P:nucleobase-containing small molecule biosynthetic process; F:nucleotide binding; F:NADH pyrophosphatase activity; F:dTTP diphosphatase activity; F:metal ion binding; C:cytoplasm | NAD(+) diphosphatase; Nucleotide diphosphatase | P:nucleoside triphosphate catabolic process; F:nucleoside triphosphate diphosphatase activity | view details | ||
| JG647394.1 | Tinospora cordifolia | citrate synthase, mitochondrial | 695 | P:GO:0005975; P:GO:0006099; P:GO:0006101; F:GO:0004108; C:GO:0005759 | P:carbohydrate metabolic process; P:tricarboxylic acid cycle; P:citrate metabolic process; F:citrate (Si)-synthase activity; C:mitochondrial matrix | Citrate synthase (unknown stereospecificity) | F:acyltransferase activity, acyl groups converted into alkyl on transfer | view details | ||
| JG650023.1 | Tinospora cordifolia | ADP-ribosylation factor 2 isoform X1 | 792 | F:GO:0003924; F:GO:0005525 | F:GTPase activity; F:GTP binding | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG647269.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 728 | P:GO:0009738; P:GO:0080163; F:GO:0004864; F:GO:0010427; F:GO:0038023; C:GO:0005634; C:GO:0005737 | P:abscisic acid-activated signaling pathway; P:regulation of protein serine/threonine phosphatase activity; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity; C:nucleus; C:cytoplasm | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG645536.1 | Tinospora cordifolia | mitochondrial phosphate carrier protein 3, mitochondrial-like | 708 | P:GO:1990547; F:GO:0005315; C:GO:0031305 | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity; C:integral component of mitochondrial inner membrane | Translocases | P:mitochondrial phosphate ion transmembrane transport; F:inorganic phosphate transmembrane transporter activity | view details | ||
| JG646097.1 | Tinospora cordifolia | 40S ribosomal protein S19-1-like | 615 | P:GO:0000028; P:GO:0006412; F:GO:0003723; F:GO:0003735; F:GO:0016740; C:GO:0022627 | P:ribosomal small subunit assembly; P:translation; F:RNA binding; F:structural constituent of ribosome; F:transferase activity; C:cytosolic small ribosomal subunit | Transferases | P:translation; F:structural constituent of ribosome; C:ribosome | view details | ||
| JG649055.1 | Tinospora cordifolia | ADP-ribosylation factor 1-like 2 | 806 | P:GO:0006886; P:GO:0016192; F:GO:0003924; F:GO:0005525 | P:intracellular protein transport; P:vesicle-mediated transport; F:GTPase activity; F:GTP binding | Nucleoside-triphosphate phosphatase | F:GTPase activity; F:GTP binding | view details | ||
| JG645922.1 | Tinospora cordifolia | thaumatin-like protein | 858 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG648221.1 | Tinospora cordifolia | Cysteine-rich repeat secretory protein 3 | 617 | P:GO:0010497; P:GO:0046739; C:GO:0009506; C:GO:0016021 | P:plasmodesmata-mediated intercellular transport; P:transport of virus in multicellular host; C:plasmodesma; C:integral component of membrane | no GO terms | view details | |||
| JG647346.1 | Tinospora cordifolia | 60S ribosomal protein L13-1 | 726 | P:GO:0006412; F:GO:0003723; F:GO:0003735; C:GO:0022625 | P:translation; F:RNA binding; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650851.1 | Tinospora cordifolia | CLK4-associating serine/arginine rich protein isoform X1 | 777 | no GO terms | view details | |||||
| JG647459.1 | Tinospora cordifolia | probable acylpyruvase FAHD1, mitochondrial | 802 | F:GO:0018773; C:GO:0005739 | F:acetylpyruvate hydrolase activity; C:mitochondrion | Acetylpyruvate hydrolase | F:catalytic activity | view details | ||
| JG647136.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 786 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648337.1 | Tinospora cordifolia | Proteinase inhibitor I3, Kunitz legume | 765 | P:GO:0010951; F:GO:0004866 | P:negative regulation of endopeptidase activity; F:endopeptidase inhibitor activity | F:endopeptidase inhibitor activity | view details | |||
| JG648548.1 | Tinospora cordifolia | thaumatin-like protein | 803 | P:GO:0006952; C:GO:0005576 | P:defense response; C:extracellular region | no GO terms | view details | |||
| JG646089.1 | Tinospora cordifolia | ribulose-phosphate 3-epimerase, cytoplasmic isoform | 746 | P:GO:0009052; P:GO:0019323; P:GO:0044262; F:GO:0004750; F:GO:0046872; C:GO:0005829 | P:pentose-phosphate shunt, non-oxidative branch; P:pentose catabolic process; P:cellular carbohydrate metabolic process; F:D-ribulose-phosphate 3-epimerase activity; F:metal ion binding; C:cytosol | Ribulose-phosphate 3-epimerase | P:carbohydrate metabolic process; P:pentose-phosphate shunt; F:D-ribulose-phosphate 3-epimerase activity; F:racemase and epimerase activity, acting on carbohydrates and derivatives | view details | ||
| JG646171.1 | Tinospora cordifolia | RING-box protein 1a | 723 | P:GO:0006511; P:GO:0016567; F:GO:0008270; F:GO:0061630; F:GO:0097602; C:GO:0005634; C:GO:0031461 | P:ubiquitin-dependent protein catabolic process; P:protein ubiquitination; F:zinc ion binding; F:ubiquitin protein ligase activity; F:cullin family protein binding; C:nucleus; C:cullin-RING ubiquitin ligase complex | Transferases | F:zinc ion binding | view details | ||
| JG650295.1 | Tinospora cordifolia | protein ABHD11 | 774 | P:GO:0006412; F:GO:0003729; F:GO:0003735; F:GO:0016787; C:GO:0005737; C:GO:0005840 | P:translation; F:mRNA binding; F:structural constituent of ribosome; F:hydrolase activity; C:cytoplasm; C:ribosome | Hydrolases | no GO terms | view details | ||
| JG646071.1 | Tinospora cordifolia | probable acetyltransferase NATA1-like | 666 | F:GO:0008080 | F:N-acetyltransferase activity | Acyltransferases | F:acetyltransferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | view details | ||
| JG648150.1 | Tinospora cordifolia | NDR1/HIN1-like protein 1 | 831 | P:GO:0098542; C:GO:0009506; C:GO:0016021; C:GO:0046658 | P:defense response to other organism; C:plasmodesma; C:integral component of membrane; C:anchored component of plasma membrane | P:defense response to other organism | view details | |||
| JG645986.1 | Tinospora cordifolia | ripening-related protein grip22 | 770 | C:GO:0005576 | C:extracellular region | no GO terms | view details | |||
| JG649591.1 | Tinospora cordifolia | cysteine proteinase inhibitor 5 | 736 | P:GO:0010466; F:GO:0030414 | P:negative regulation of peptidase activity; F:peptidase inhibitor activity | F:cysteine-type endopeptidase inhibitor activity | view details | |||
| JG650108.1 | Tinospora cordifolia | probable calcium-binding protein CML36 | 791 | F:GO:0005509 | F:calcium ion binding | F:calcium ion binding | view details | |||
| JG648537.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 638 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG650596.1 | Tinospora cordifolia | pathogenesis-related protein PR-4-like | 670 | P:GO:0042742; P:GO:0050832; P:GO:0090501; F:GO:0004540 | P:defense response to bacterium; P:defense response to fungus; P:RNA phosphodiester bond hydrolysis; F:ribonuclease activity | Acting on ester bonds | P:defense response; P:defense response to bacterium; P:defense response to fungus; F:ribonuclease activity | view details | ||
| JG649627.1 | Tinospora cordifolia | transmembrane protein | 808 | C:GO:0016021 | C:integral component of membrane | no GO terms | view details | |||
| JG650322.1 | Tinospora cordifolia | glutathione reductase, cytosolic-like | 743 | P:GO:0006749; P:GO:0045454; P:GO:0098869; F:GO:0004362; F:GO:0004791; F:GO:0050660; F:GO:0050661; C:GO:0005737 | P:glutathione metabolic process; P:cell redox homeostasis; P:cellular oxidant detoxification; F:glutathione-disulfide reductase (NADPH) activity; F:thioredoxin-disulfide reductase activity; F:flavin adenine dinucleotide binding; F:NADP binding; C:cytoplasm | Glutathione-disulfide reductase; Thioredoxin-disulfide reductase; Protein-disulfide reductase | F:oxidoreductase activity | view details | ||
| JG650381.1 | Tinospora cordifolia | 40S ribosomal protein S24-1 | 766 | P:GO:0006412; F:GO:0003729; F:GO:0003735; C:GO:0005829; C:GO:0005840 | P:translation; F:mRNA binding; F:structural constituent of ribosome; C:cytosol; C:ribosome | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG650618.1 | Tinospora cordifolia | thaumatin-like protein | 793 | no GO terms | view details | |||||
| JG648411.1 | Tinospora cordifolia | Mannose-binding lectin | 665 | F:GO:0005537; F:GO:0030246; C:GO:0016020; C:GO:0016021 | F:mannose binding; F:carbohydrate binding; C:membrane; C:integral component of membrane | no GO terms | view details | |||
| JG647791.1 | Tinospora cordifolia | thioredoxin M3, chloroplastic | 781 | P:GO:0006662; P:GO:0043085; F:GO:0008047; F:GO:0015035; F:GO:0016671; C:GO:0005737 | P:glycerol ether metabolic process; P:positive regulation of catalytic activity; F:enzyme activator activity; F:protein-disulfide reductase activity; F:oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor; C:cytoplasm | Acting on a sulfur group of donors | no GO terms | view details | ||
| JG647012.1 | Tinospora cordifolia | hypothetical protein IFM89_025060 | 835 | no GO terms | view details | |||||
| JG646404.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 808 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG647652.1 | Tinospora cordifolia | Bet v I domain | 757 | F:GO:0016829; C:GO:0016020 | F:lyase activity; C:membrane | Lyases | P:defense response | view details | ||
| JG648543.1 | Tinospora cordifolia | epidermis-specific secreted glycoprotein EP1-like | 815 | P:GO:0048544; C:GO:0016021 | P:recognition of pollen; C:integral component of membrane | no GO terms | view details | |||
| JG649280.1 | Tinospora cordifolia | major allergen Pru ar 1-like | 716 | P:GO:0050790; P:GO:0050794; P:GO:0050896; C:GO:0005622 | P:regulation of catalytic activity; P:regulation of cellular process; P:response to stimulus; C:intracellular anatomical structure | P:defense response; P:abscisic acid-activated signaling pathway; F:protein phosphatase inhibitor activity; F:abscisic acid binding; F:signaling receptor activity | view details | |||
| JG648469.1 | Tinospora cordifolia | thaumatin-like protein | 862 | no GO terms | view details | |||||
| JG650348.1 | Tinospora cordifolia | 60S ribosomal protein L18a | 749 | P:GO:0006412; F:GO:0003735; C:GO:0022625 | P:translation; F:structural constituent of ribosome; C:cytosolic large ribosomal subunit | P:translation; F:structural constituent of ribosome; C:ribosome | view details | |||
| JG648683.1 | Tinospora cordifolia | ubiquitin-conjugating enzyme E2 variant 1D | 822 | P:GO:0006301; P:GO:0070534; C:GO:0005634 | P:postreplication repair; P:protein K63-linked ubiquitination; C:nucleus | no GO terms | view details | |||
| MS688122.1 | Tinospora cordifolia | stemmadenine O-acetyltransferase-like | 1792 | F:GO:0016740; F:GO:0016747 | F:transferase activity; F:acyltransferase activity, transferring groups other than amino-acyl groups | no GO terms | view details | |||
| JG646644.1 | Tinospora cordifolia | corytuberine synthase | 795 | P:GO:0002238; P:GO:0009699; P:GO:1901012; F:GO:0005506; F:GO:0020037; F:GO:0050593; F:GO:0102963; C:GO:0005789; C:GO:0016021 | P:response to molecule of fungal origin; P:phenylpropanoid biosynthetic process; P:(S)-reticuline biosynthetic process; F:iron ion binding; F:heme binding; F:N-methylcoclaurine 3-monooxygenase activity; F:(S)-corytuberine synthase activity; C:endoplasmic reticulum membrane; C:integral component of membrane | (S)-corytuberine synthase; N-methylcoclaurine 3-monooxygenase; Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG645813.1 | Tinospora cordifolia | probable tetraacyldisaccharide 4-kinase, mitochondrial | 831 | P:GO:0009245; P:GO:0016310; F:GO:0005524; F:GO:0009029; C:GO:0016020 | P:lipid A biosynthetic process; P:phosphorylation; F:ATP binding; F:tetraacyldisaccharide 4-kinase activity; C:membrane | Tetraacyldisaccharide 4-kinase | P:lipid A biosynthetic process; F:ATP binding; F:tetraacyldisaccharide 4-kinase activity | view details | ||
| JG649571.1 | Tinospora cordifolia | pseudouridine-5-phosphate glycosidase | 749 | P:GO:0001522; F:GO:0004730; F:GO:0016798; F:GO:0046872 | P:pseudouridine synthesis; F:pseudouridylate synthase activity; F:hydrolase activity, acting on glycosyl bonds; F:metal ion binding | Glycosylases; Pseudouridylate synthase | F:pseudouridylate synthase activity; F:hydrolase activity, acting on glycosyl bonds | view details | ||
| JG649626.1 | Tinospora cordifolia | UMP-CMP kinase 3-like isoform X1 | 624 | P:GO:0006207; P:GO:0006225; P:GO:0016310; P:GO:0046705; P:GO:0046940; F:GO:0005524; F:GO:0033862; F:GO:0036430; F:GO:0036431; C:GO:0005634; C:GO:0009507 | P:de novo pyrimidine nucleobase biosynthetic process; P:UDP biosynthetic process; P:phosphorylation; P:CDP biosynthetic process; P:nucleoside monophosphate phosphorylation; F:ATP binding; F:UMP kinase activity; F:CMP kinase activity; F:dCMP kinase activity; C:nucleus; C:chloroplast | Nucleoside-phosphate kinase; UMP kinase; UMP/CMP kinase | P:nucleobase-containing compound metabolic process; F:ATP binding; F:nucleobase-containing compound kinase activity | view details | ||
| JG645729.1 | Tinospora cordifolia | exonuclease V, chloroplastic | 839 | P:GO:0036297; F:GO:0045145; C:GO:0005634 | P:interstrand cross-link repair; F:single-stranded DNA 5-3 exodeoxyribonuclease activity; C:nucleus | Acting on ester bonds; Acting on ester bonds | F:single-stranded DNA 5-3 exodeoxyribonuclease activity | view details | ||
| JG646337.1 | Tinospora cordifolia | serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B kappa isoform-like | 780 | P:GO:0007165; P:GO:0050790; F:GO:0019888; C:GO:0000159; C:GO:0005737 | P:signal transduction; P:regulation of catalytic activity; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex; C:cytoplasm | P:signal transduction; F:protein phosphatase regulator activity; C:protein phosphatase type 2A complex | view details | |||
| JG647484.1 | Tinospora cordifolia | DNA-directed RNA polymerase II subunit 4 | 828 | P:GO:0000288; P:GO:0006367; P:GO:0031990; P:GO:0034402; P:GO:0045948; F:GO:0000166; F:GO:0003899; F:GO:0031369; C:GO:0000932; C:GO:0005665 | P:nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay; P:transcription initiation at RNA polymerase II promoter; P:mRNA export from nucleus in response to heat stress; P:recruitment of 3-end processing factors to RNA polymerase II holoenzyme complex; P:positive regulation of translational initiation; F:nucleotide binding; F:DNA-directed 5-3 RNA polymerase activity; F:translation initiation factor binding; C:P-body; C:RNA polymerase II, core complex | DNA-directed RNA polymerase | P:DNA-templated transcription initiation; P:cellular metabolic process; F:nucleotide binding; C:RNA polymerase complex | view details | ||
| JG646142.1 | Tinospora cordifolia | probable (S)-N-methylcoclaurine 3-hydroxylase isozyme 2 | 747 | P:GO:0019438; P:GO:1901362; F:GO:0004497; F:GO:0005506; F:GO:0016705; F:GO:0020037 | P:aromatic compound biosynthetic process; P:organic cyclic compound biosynthetic process; F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | Acting on paired donors, with incorporation or reduction of molecular oxygen. The oxygen incorporated need not be derived from O(2) | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details | ||
| JG650701.1 | Tinospora cordifolia | phloretin 4-O-glucosyltransferase-like | 749 | F:GO:0016757 | F:glycosyltransferase activity | Glycosyltransferases | F:UDP-glycosyltransferase activity | view details | ||
| JG649746.1 | Tinospora cordifolia | Expression of the gene is downregulated in the presence of paraquat, putative | 535 | P:GO:0006979; P:GO:0032774; F:GO:0003899 | P:response to oxidative stress; P:RNA biosynthetic process; F:DNA-directed 5-3 RNA polymerase activity | no GO terms | view details | |||
| JH648119.1 | Tinospora cordifolia | Glutathione S-transferase | 426 | P:GO:0006749; F:GO:0004364; C:GO:0005737 | P:glutathione metabolic process; F:glutathione transferase activity; C:cytoplasm | Glutathione transferase | F:glutathione transferase activity | view details | ||
| JI646017.1 | Tinospora cordifolia | cytochrome P450 81E8-like | 796 | P:GO:0009058; F:GO:0005488; F:GO:0016491 | P:biosynthetic process; F:binding; F:oxidoreductase activity | Oxidoreductases | F:monooxygenase activity; F:iron ion binding; F:oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; F:heme binding | view details |
lncRNA Information
Gene Information
| Gene ID | Species Name | Symbol | Aliases | Description | Start position | End position | Orientation | Genomic Context |
|---|---|---|---|---|---|---|---|---|
| 40136794 | Tinospora cordifolia | clpP | FDY42_pgp041 | ATP-dependent Clp protease proteolytic subunit | 69465 | 71552 | minus | |
| 40136790 | Tinospora cordifolia | rps7 | FDY42_pgp020 | ribosomal protein S7 | 96181 | 96648 | minus | |
| 40136789 | Tinospora cordifolia | ycf15 | FDY42_pgp019 | Ycf15 | 98293 | 98691 | minus | |
| 40136786 | Tinospora cordifolia | ndhI | FDY42_pgp011 | NADH dehydrogenase subunit I | 116374 | 116916 | minus | |
| 40136784 | Tinospora cordifolia | ndhA | FDY42_pgp010 | NADH dehydrogenase subunit 1 | 116999 | 119156 | minus | |
| 40136783 | Tinospora cordifolia | rps7 | FDY42_pgp005 | ribosomal protein S7 | 138181 | 138648 | plus | |
| 40136782 | Tinospora cordifolia | ndhG | FDY42_pgp012 | NADH dehydrogenase subunit 6 | 115423 | 115953 | minus | |
| 40136780 | Tinospora cordifolia | psaC | FDY42_pgp014 | photosystem I subunit VII | 114361 | 114606 | minus | |
| 40136778 | Tinospora cordifolia | ndhE | FDY42_pgp013 | NADH dehydrogenase subunit 4L | 114871 | 115176 | minus | |
| 40136777 | Tinospora cordifolia | rpl2 | FDY42_pgp001 | ribosomal protein L2 | 149404 | 150887 | plus | |
| 40136776 | Tinospora cordifolia | ndhB | FDY42_pgp004 | NADH dehydrogenase subunit 2 | 138978 | 141204 | plus | |
| 40136775 | Tinospora cordifolia | ndhH | FDY42_pgp009 | NADH dehydrogenase subunit 7 | 119158 | 120339 | minus | |
| 40136774 | Tinospora cordifolia | ndhD | FDY42_pgp015 | NADH dehydrogenase subunit 4 | 112735 | 114255 | minus | |
| 40136773 | Tinospora cordifolia | rps15 | FDY42_pgp008 | ribosomal protein S15 | 120456 | 120728 | minus | |
| 40136772 | Tinospora cordifolia | rpl32 | FDY42_pgp017 | ribosomal protein L32 | 110956 | 111129 | plus | |
| 40136764 | Tinospora cordifolia | rpl20 | FDY42_pgp044 | ribosomal protein L20 | 68134 | 68493 | minus | |
| 40136762 | Tinospora cordifolia | psbH | FDY42_pgp037 | photosystem II protein H | 74133 | 74354 | plus | |
| 40136761 | Tinospora cordifolia | petG | FDY42_pgp048 | cytochrome b6/f complex subunit V | 65721 | 65834 | plus | |
| 40136760 | Tinospora cordifolia | psbI | FDY42_pgp083 | photosystem II protein I | 7461 | 7571 | plus | |
| 40136759 | Tinospora cordifolia | rps12 | FDY42_pgp043 | ribosomal protein S12 | 69276 | 69389 | minus | |
| 40136759 | Tinospora cordifolia | rps12 | FDY42_pgp043 | ribosomal protein S12 | 137334 | 138127 | plus | |
| 40136758 | Tinospora cordifolia | rpl2 | FDY42_pgp024 | ribosomal protein L2 | 83967 | 85438 | minus | |
| 40136757 | Tinospora cordifolia | psaB | FDY42_pgp068 | photosystem I P700 chlorophyll a apoprotein A2 | 36556 | 38760 | minus | |
| 40136756 | Tinospora cordifolia | rps19 | FDY42_pgp025 | ribosomal protein S19 | 83591 | 83869 | minus | |
| 40136755 | Tinospora cordifolia | rps3 | FDY42_pgp027 | ribosomal protein S3 | 82363 | 83019 | minus | |
| 40136754 | Tinospora cordifolia | psbB | FDY42_pgp055 | photosystem II 47 kDa protein | 60483 | 62009 | plus | |
| 40136753 | Tinospora cordifolia | rpoA | FDY42_pgp034 | RNA polymerase alpha subunit | 77535 | 78554 | minus | |
| 40136752 | Tinospora cordifolia | rps2 | FDY42_pgp078 | ribosomal protein S2 | 14829 | 15539 | minus | |
| 40136751 | Tinospora cordifolia | rpoC2 | FDY42_pgp077 | RNA polymerase beta' subunit | 15628 | 19869 | plus | |
| 40136750 | Tinospora cordifolia | psbJ | FDY42_pgp053 | photosystem II protein J | 63658 | 63780 | minus | |
| 40136749 | Tinospora cordifolia | psbC | FDY42_pgp071 | photosystem II 44 kDa protein | 33131 | 34516 | plus | |
| 40136748 | Tinospora cordifolia | psaI | FDY42_pgp057 | photosystem I subunit VIII | 58717 | 58827 | plus | |
| 40136747 | Tinospora cordifolia | accD | FDY42_pgp058 | acetyl-CoA carboxylase beta subunit | 56486 | 57979 | plus | |
| 40136746 | Tinospora cordifolia | rbcL | FDY42_pgp059 | ribulose-1 | 54310 | 55758 | plus | |
| 40136745 | Tinospora cordifolia | ycf3 | FDY42_pgp066 | photosystem I assembly protein Ycf3 | 41663 | 43666 | minus | |
| 40136744 | Tinospora cordifolia | psbB | FDY42_pgp040 | photosystem II 47 kDa protein | 72022 | 73548 | plus | |
| 40136743 | Tinospora cordifolia | ycf4 | FDY42_pgp056 | photosystem I assembly protein Ycf4 | 59217 | 59771 | plus | |
| 40136742 | Tinospora cordifolia | psbN | FDY42_pgp038 | photosystem II protein N | 73907 | 74038 | minus | |
| 40136741 | Tinospora cordifolia | rpl36 | FDY42_pgp032 | ribosomal protein L36 | 79156 | 79269 | minus | |
| 40136739 | Tinospora cordifolia | rpl33 | FDY42_pgp046 | ribosomal protein L33 | 67228 | 67428 | plus | |
| 40136737 | Tinospora cordifolia | rps18 | FDY42_pgp045 | ribosomal protein S18 | 67592 | 67897 | plus | |
| 40136735 | Tinospora cordifolia | rpl16 | FDY42_pgp028 | ribosomal protein L16 | 80772 | 81176 | minus | |
| 40136733 | Tinospora cordifolia | rpl14 | FDY42_pgp029 | ribosomal protein L14 | 80258 | 80626 | minus | |
| 40136731 | Tinospora cordifolia | petD | FDY42_pgp035 | cytochrome b6/f complex subunit IV | 76801 | 77346 | plus | |
| 40136730 | Tinospora cordifolia | rps11 | FDY42_pgp033 | ribosomal protein S11 | 78620 | 79036 | minus | |
| 40136728 | Tinospora cordifolia | rpoB | FDY42_pgp075 | RNA polymerase beta subunit | 22886 | 26098 | minus | |
| 40136726 | Tinospora cordifolia | psbM | FDY42_pgp073 | photosystem II protein M | 28663 | 28779 | plus | |
| 40136723 | Tinospora cordifolia | psbF | FDY42_pgp051 | photosystem II protein VI | 64042 | 64161 | minus | |
| 40136722 | Tinospora cordifolia | psbE | FDY42_pgp050 | photosystem II protein V | 64171 | 64422 | minus | |
| 40136721 | Tinospora cordifolia | atpI | FDY42_pgp079 | ATP synthase CF0 A subunit | 13862 | 14605 | minus | |
| 40136720 | Tinospora cordifolia | petB | FDY42_pgp036 | cytochrome b6 | 75245 | 75931 | plus | |
| 40136719 | Tinospora cordifolia | rps8 | FDY42_pgp030 | ribosomal protein S8 | 79667 | 80065 | minus | |
| 40136718 | Tinospora cordifolia | petL | FDY42_pgp049 | cytochrome b6/f complex subunit VI | 65442 | 65537 | plus | |
| 40136716 | Tinospora cordifolia | atpH | FDY42_pgp080 | ATP synthase CF0 C subunit | 12600 | 12845 | minus | |
| 40136715 | Tinospora cordifolia | atpB | FDY42_pgp060 | ATP synthase CF1 beta subunit | 52004 | 53500 | minus | |
| 40136714 | Tinospora cordifolia | rpl23 | FDY42_pgp023 | ribosomal protein L23 | 85445 | 85726 | minus | |
| 40136713 | Tinospora cordifolia | ndhK | FDY42_pgp063 | NADH dehydrogenase subunit K | 49031 | 49708 | minus | |
| 40136712 | Tinospora cordifolia | rpl22 | FDY42_pgp026 | ribosomal protein L22 | 83004 | 83528 | minus | |
| 40136711 | Tinospora cordifolia | ndhC | FDY42_pgp062 | NADH dehydrogenase subunit 3 | 49767 | 50129 | minus | |
| 40136710 | Tinospora cordifolia | matK | FDY42_pgp086 | maturase K | 1864 | 3393 | minus | |
| 40136708 | Tinospora cordifolia | atpF | FDY42_pgp081 | ATP synthase CF0 B subunit | 11251 | 12499 | minus | |
| 40136707 | Tinospora cordifolia | psbT | FDY42_pgp039 | photosystem II protein T | 73741 | 73848 | plus | |
| 40136706 | Tinospora cordifolia | petA | FDY42_pgp054 | cytochrome f | 62233 | 63195 | plus | |
| 40136705 | Tinospora cordifolia | psaA | FDY42_pgp067 | photosystem I P700 chlorophyll a apoprotein A1 | 38786 | 41038 | minus | |
| 40136703 | Tinospora cordifolia | infA | FDY42_pgp031 | translation initiation factor 1 | 79375 | 79608 | minus | |
| 40136702 | Tinospora cordifolia | psbL | FDY42_pgp052 | photosystem II protein L | 63900 | 64016 | minus | |
| 40136699 | Tinospora cordifolia | atpE | FDY42_pgp061 | ATP synthase CF1 epsilon subunit | 51606 | 52007 | minus | |
| 40136698 | Tinospora cordifolia | ndhJ | FDY42_pgp064 | NADH dehydrogenase subunit J | 48407 | 48883 | minus | |
| 40136697 | Tinospora cordifolia | rps4 | FDY42_pgp065 | ribosomal protein S4 | 44942 | 45547 | minus | |
| 40136695 | Tinospora cordifolia | psbD | FDY42_pgp072 | photosystem II protein D2 | 32161 | 33147 | plus | |
| 40136694 | Tinospora cordifolia | petN | FDY42_pgp074 | cytochrome b6/f complex subunit VIII | 27822 | 27911 | plus | |
| 40136693 | Tinospora cordifolia | psbA | FDY42_pgp087 | photosystem II protein D1 | 305 | 1366 | minus | |
| 40136692 | Tinospora cordifolia | psbK | FDY42_pgp084 | photosystem II protein K | 6986 | 7171 | plus | |
| 40136687 | Tinospora cordifolia | psbZ | FDY42_pgp070 | photosystem II protein Z | 35147 | 35335 | plus | |
| 40136686 | Tinospora cordifolia | rps14 | FDY42_pgp069 | ribosomal protein S14 | 36110 | 36412 | minus | |
| 40136683 | Tinospora cordifolia | atpA | FDY42_pgp082 | ATP synthase CF1 alpha subunit | 9647 | 11170 | minus | |
| 40136682 | Tinospora cordifolia | psaJ | FDY42_pgp047 | photosystem I subunit IX | 66612 | 66746 | plus | |
| 40136681 | Tinospora cordifolia | ycf2 | FDY42_pgp022 | Ycf2 | 87812 | 92554 | plus | |
| 40136680 | Tinospora cordifolia | ndhB | FDY42_pgp021 | NADH dehydrogenase subunit 2 | 93625 | 95851 | minus | |
| 40136679 | Tinospora cordifolia | ndhF | FDY42_pgp018 | NADH dehydrogenase subunit 5 | 108045 | 110252 | minus | |
| 40136677 | Tinospora cordifolia | ycf15 | FDY42_pgp006 | Ycf15 | 136134 | 136328 | plus | |
| 40136676 | Tinospora cordifolia | rps12 | FDY42_pgp042 | ribosomal protein S12 | 69276 | 69389 | minus | |
| 40136676 | Tinospora cordifolia | rps12 | FDY42_pgp042 | ribosomal protein S12 | 96702 | 97495 | minus | |
| 40136675 | Tinospora cordifolia | rpl23 | FDY42_pgp002 | ribosomal protein L23 | 149104 | 149385 | plus | |
| 40136674 | Tinospora cordifolia | ycf2 | FDY42_pgp003 | Ycf2 | 142275 | 148799 | minus | |
| 40136672 | Tinospora cordifolia | ccsA | FDY42_pgp016 | cytochrome c biogenesis protein | 111537 | 112508 | plus | |
| 40136779 | Tinospora cordifolia | ycf1 | FDY42_pgp007 | pseudo | 121180 | 126758 | minus | |
| 40136771 | Tinospora cordifolia | rrn4.5S | FDY42_pgr003 | 4.5S ribosomal RNA | 128438 | 128540 | plus | |
| 40136769 | Tinospora cordifolia | rrn23S | FDY42_pgr002 | 23S ribosomal RNA | 128640 | 132053 | minus | |
| 40136768 | Tinospora cordifolia | rrn5S | FDY42_pgr005 | 5S ribosomal RNA | 106610 | 106740 | plus | |
| 40136766 | Tinospora cordifolia | rrn16S | FDY42_pgr001 | 16S ribosomal RNA | 133845 | 135335 | plus | |
| 40136765 | Tinospora cordifolia | rrn4.5S | FDY42_pgr006 | 4.5S ribosomal RNA | 106285 | 106387 | minus | |
| 40136709 | Tinospora cordifolia | rps16 | FDY42_pgp085 | pseudo | 4516 | 4761 | minus | |
| 40136700 | Tinospora cordifolia | rrn23S | FDY42_pgr007 | 23S ribosomal RNA | 102771 | 106185 | plus | |
| 40136678 | Tinospora cordifolia | rrn5S | FDY42_pgr004 | 5S ribosomal RNA | 128085 | 128215 | minus | |
| 40136793 | Tinospora cordifolia | trnH-GTG | FDY42_pgt001 | tRNA | 59 | 132 | minus | |
| 40136792 | Tinospora cordifolia | trnL-CAA | FDY42_pgt020 | tRNA | 92990 | 93070 | minus | |
| 40136791 | Tinospora cordifolia | trnV-GAC | FDY42_pgt021 | tRNA | 99191 | 99262 | plus | |
| 40136788 | Tinospora cordifolia | trnI-CAT | FDY42_pgt029 | tRNA | 148868 | 148941 | plus | |
| 40136787 | Tinospora cordifolia | trnN-GTT | FDY42_pgt025 | tRNA | 127095 | 127166 | plus | |
| 40136785 | Tinospora cordifolia | trnL-TAG | FDY42_pgt024 | tRNA | 111434 | 111513 | plus | |
| 40136781 | Tinospora cordifolia | trnL-CAA | FDY42_pgt028 | tRNA | 141759 | 141839 | plus | |
| 40136770 | Tinospora cordifolia | trnR-ACG | FDY42_pgt022 | tRNA | 106973 | 107046 | plus | |
| 40136767 | Tinospora cordifolia | trnV-GAC | FDY42_pgt027 | tRNA | 135563 | 135634 | minus | |
| 40136763 | Tinospora cordifolia | trnS-GGA | FDY42_pgt013 | tRNA | 44560 | 44649 | plus | |
| 40136740 | Tinospora cordifolia | trnD-GTC | FDY42_pgt006 | tRNA | 29172 | 29245 | minus | |
| 40136738 | Tinospora cordifolia | trnG-GCC | FDY42_pgt011 | tRNA | 35643 | 35713 | plus | |
| 40136736 | Tinospora cordifolia | trnQ-TTG | FDY42_pgt002 | tRNA | 6559 | 6630 | minus | |
| 40136734 | Tinospora cordifolia | trnM-CAT | FDY42_pgt016 | tRNA | 51295 | 51367 | plus | |
| 40136732 | Tinospora cordifolia | trnT-TGT | FDY42_pgt014 | tRNA | 45923 | 45995 | minus | |
| 40136729 | Tinospora cordifolia | trnS-TGA | FDY42_pgt010 | tRNA | 34712 | 34804 | minus | |
| 40136727 | Tinospora cordifolia | trnC-GCA | FDY42_pgt005 | tRNA | 27310 | 27380 | plus | |
| 40136725 | Tinospora cordifolia | trnW-CCA | FDY42_pgt017 | tRNA | 65980 | 66053 | minus | |
| 40136724 | Tinospora cordifolia | trnS-GCT | FDY42_pgt003 | tRNA | 7712 | 7799 | minus | |
| 40136717 | Tinospora cordifolia | trnI-CAT | FDY42_pgt019 | tRNA | 85889 | 85962 | minus | |
| 40136701 | Tinospora cordifolia | trnN-GTT | FDY42_pgt023 | tRNA | 107664 | 107735 | minus | |
| 40136696 | Tinospora cordifolia | trnfM-CAT | FDY42_pgt012 | tRNA | 35875 | 35948 | minus | |
| 40136691 | Tinospora cordifolia | trnR-TCT | FDY42_pgt004 | tRNA | 9458 | 9529 | plus | |
| 40136690 | Tinospora cordifolia | trnY-GTA | FDY42_pgt007 | tRNA | 29721 | 29804 | minus | |
| 40136689 | Tinospora cordifolia | trnE-TTC | FDY42_pgt008 | tRNA | 29864 | 29936 | minus | |
| 40136688 | Tinospora cordifolia | trnT-GGT | FDY42_pgt009 | tRNA | 30667 | 30738 | plus | |
| 40136685 | Tinospora cordifolia | trnF-GAA | FDY42_pgt015 | tRNA | 47598 | 47670 | plus | |
| 40136684 | Tinospora cordifolia | trnP-TGG | FDY42_pgt018 | tRNA | 66225 | 66298 | minus | |
| 40136704 | Tinospora cordifolia | rpoC1 | FDY42_pgp076 | RNA polymerase beta subunit | 20026 | 22859 | minus | |
| 40136704 | Tinospora cordifolia | rpoC1 | FDY42_pgp076 | RNA polymerase beta subunit | 20026 | 22859 | minus |
miRNA Information
| Acc. No. | Species Name | Sequence length | Total pre-miRNA | Sequence |
|---|---|---|---|---|
| JG645598.1 | Tinospora cordifolia | 700 | 7 Details |
|
| JG649774.1 | Tinospora cordifolia | 745 | 7 Details |
|
| JG646035.1 | Tinospora cordifolia | 852 | 5 Details |
|
| JG650192.1 | Tinospora cordifolia | 747 | 6 Details |
|
| JG648729.1 | Tinospora cordifolia | 864 | 9 Details |
|
| JG649100.1 | Tinospora cordifolia | 815 | 7 Details |
|
| JG646358.1 | Tinospora cordifolia | 740 | 5 Details |
|
| JG649164.1 | Tinospora cordifolia | 863 | 4 Details |
|
| JG647116.1 | Tinospora cordifolia | 846 | 8 Details |
|
| JG648441.1 | Tinospora cordifolia | 881 | 7 Details |
|
| JG647165.1 | Tinospora cordifolia | 806 | 4 Details |
|
| JG648539.1 | Tinospora cordifolia | 797 | 7 Details |
|
| JG647274.1 | Tinospora cordifolia | 832 | 6 Details |
|
| JG649628.1 | Tinospora cordifolia | 762 | 6 Details |
|
| JG645532.1 | Tinospora cordifolia | 739 | 5 Details |
|
| JG650838.1 | Tinospora cordifolia | 826 | 7 Details |
|
| JG646080.1 | Tinospora cordifolia | 746 | 7 Details |
|
| JG649647.1 | Tinospora cordifolia | 784 | 7 Details |
|
| JG647335.1 | Tinospora cordifolia | 776 | 6 Details |
|
| JG648312.1 | Tinospora cordifolia | 772 | 8 Details |
|
| JG649846.1 | Tinospora cordifolia | 817 | 7 Details |
|
| JG649949.1 | Tinospora cordifolia | 607 | 6 Details |
|
| JG647190.1 | Tinospora cordifolia | 799 | 8 Details |
|
| JG647468.1 | Tinospora cordifolia | 737 | 6 Details |
|
| JG650926.1 | Tinospora cordifolia | 847 | 8 Details |
|
| JG647466.1 | Tinospora cordifolia | 783 | 7 Details |
|
| JG646451.1 | Tinospora cordifolia | 796 | 8 Details |
|
| JG650947.1 | Tinospora cordifolia | 760 | 7 Details |
|
| JG649067.1 | Tinospora cordifolia | 793 | 7 Details |
|
| JG649258.1 | Tinospora cordifolia | 641 | 5 Details |
|
| JG648496.1 | Tinospora cordifolia | 833 | 8 Details |
|
| JG649251.1 | Tinospora cordifolia | 741 | 8 Details |
|
| JG647728.1 | Tinospora cordifolia | 756 | 7 Details |
|
| JG646629.1 | Tinospora cordifolia | 797 | 5 Details |
|
| JG646184.1 | Tinospora cordifolia | 603 | 6 Details |
|
| JG650247.1 | Tinospora cordifolia | 825 | 7 Details |
|
| JG649141.1 | Tinospora cordifolia | 825 | 6 Details |
|
| JG650047.1 | Tinospora cordifolia | 778 | 5 Details |
|
| JG646285.1 | Tinospora cordifolia | 820 | 8 Details |
|
| JG648200.1 | Tinospora cordifolia | 821 | 6 Details |
|
| JG647174.1 | Tinospora cordifolia | 797 | 7 Details |
|
| JG650798.1 | Tinospora cordifolia | 844 | 5 Details |
|
| JG646867.1 | Tinospora cordifolia | 672 | 6 Details |
|
| JG646748.1 | Tinospora cordifolia | 793 | 7 Details |
|
| JG650097.1 | Tinospora cordifolia | 700 | 7 Details |
|
| JG649285.1 | Tinospora cordifolia | 710 | 7 Details |
|
| JG646716.1 | Tinospora cordifolia | 786 | 9 Details |
|
| JG650603.1 | Tinospora cordifolia | 774 | 7 Details |
|
| JG648953.1 | Tinospora cordifolia | 802 | 7 Details |
|
| JG647702.1 | Tinospora cordifolia | 831 | 8 Details |
|
| JG648925.1 | Tinospora cordifolia | 798 | 8 Details |
|
| JG646353.1 | Tinospora cordifolia | 738 | 6 Details |
|
| JG649440.1 | Tinospora cordifolia | 806 | 5 Details |
|
| JG648410.1 | Tinospora cordifolia | 806 | 2 Details |
|
| JG646545.1 | Tinospora cordifolia | 675 | 7 Details |
|
| JG650734.1 | Tinospora cordifolia | 769 | 6 Details |
|
| JG647004.1 | Tinospora cordifolia | 845 | 8 Details |
|
| JG648868.1 | Tinospora cordifolia | 778 | 5 Details |
|
| JG646124.1 | Tinospora cordifolia | 754 | 6 Details |
|
| JG649822.1 | Tinospora cordifolia | 721 | 4 Details |
|
| JG649752.1 | Tinospora cordifolia | 567 | 3 Details |
|
| JG646381.1 | Tinospora cordifolia | 807 | 8 Details |
|
| JG647226.1 | Tinospora cordifolia | 801 | 5 Details |
|
| JG648097.1 | Tinospora cordifolia | 808 | 7 Details |
|
| JG645811.1 | Tinospora cordifolia | 829 | 8 Details |
|
| JG650996.1 | Tinospora cordifolia | 758 | 5 Details |
|
| JG648012.1 | Tinospora cordifolia | 863 | 9 Details |
|
| JG649526.1 | Tinospora cordifolia | 801 | 9 Details |
|
| JG650955.1 | Tinospora cordifolia | 699 | 6 Details |
|
| JG649048.1 | Tinospora cordifolia | 850 | 6 Details |
|
| JG650188.1 | Tinospora cordifolia | 842 | 6 Details |
|
| JG650334.1 | Tinospora cordifolia | 806 | 7 Details |
|
| JG646242.1 | Tinospora cordifolia | 656 | 5 Details |
|
| JG645565.1 | Tinospora cordifolia | 678 | 3 Details |
|
| JG646345.1 | Tinospora cordifolia | 821 | 9 Details |
|
| JG649434.1 | Tinospora cordifolia | 805 | 7 Details |
|
| JG649665.1 | Tinospora cordifolia | 855 | 8 Details |
|
| JG650563.1 | Tinospora cordifolia | 843 | 8 Details |
|
| JG647223.1 | Tinospora cordifolia | 747 | 2 Details |
|
| JG650864.1 | Tinospora cordifolia | 767 | 7 Details |
|
| JG647789.1 | Tinospora cordifolia | 780 | 9 Details |
|
| JG646119.1 | Tinospora cordifolia | 805 | 5 Details |
|
| JG646357.1 | Tinospora cordifolia | 850 | 5 Details |
|
| JG646999.1 | Tinospora cordifolia | 769 | 7 Details |
|
| JG649187.1 | Tinospora cordifolia | 673 | 5 Details |
|
| JG647923.1 | Tinospora cordifolia | 828 | 6 Details |
|
| JG645630.1 | Tinospora cordifolia | 760 | 8 Details |
|
| JG648834.1 | Tinospora cordifolia | 854 | 9 Details |
|
| JG646292.1 | Tinospora cordifolia | 774 | 8 Details |
|
| JG650107.1 | Tinospora cordifolia | 845 | 5 Details |
|
| JG647246.1 | Tinospora cordifolia | 807 | 8 Details |
|
| JG647553.1 | Tinospora cordifolia | 760 | 5 Details |
|
| JG647372.1 | Tinospora cordifolia | 804 | 7 Details |
|
| JG649755.1 | Tinospora cordifolia | 698 | 6 Details |
|
| JG648687.1 | Tinospora cordifolia | 776 | 5 Details |
|
| JG648933.1 | Tinospora cordifolia | 727 | 4 Details |
|
| JG648563.1 | Tinospora cordifolia | 667 | 5 Details |
|
| JG647187.1 | Tinospora cordifolia | 796 | 8 Details |
|
| JG646207.1 | Tinospora cordifolia | 657 | 6 Details |
|
| JG645826.1 | Tinospora cordifolia | 821 | 6 Details |
|
| JG648394.1 | Tinospora cordifolia | 788 | 4 Details |
|
| JG646339.1 | Tinospora cordifolia | 760 | 7 Details |
|
| JG647314.1 | Tinospora cordifolia | 756 | 6 Details |
|
| JG648850.1 | Tinospora cordifolia | 760 | 6 Details |
|
| JG645674.1 | Tinospora cordifolia | 769 | 5 Details |
|
| JG649726.1 | Tinospora cordifolia | 643 | 7 Details |
|
| JG650184.1 | Tinospora cordifolia | 714 | 8 Details |
|
| JG648832.1 | Tinospora cordifolia | 844 | 8 Details |
|
| JG648070.1 | Tinospora cordifolia | 841 | 6 Details |
|
| JG649809.1 | Tinospora cordifolia | 712 | 5 Details |
|
| JG649139.1 | Tinospora cordifolia | 767 | 4 Details |
|
| JG649969.1 | Tinospora cordifolia | 536 | 4 Details |
|
| JG646238.1 | Tinospora cordifolia | 432 | 3 Details |
|
| JG646022.1 | Tinospora cordifolia | 823 | 6 Details |
|
| JG646485.1 | Tinospora cordifolia | 833 | 6 Details |
|
| JG650811.1 | Tinospora cordifolia | 805 | 6 Details |
|
| JG647391.1 | Tinospora cordifolia | 725 | 7 Details |
|
| JG648354.1 | Tinospora cordifolia | 824 | 7 Details |
|
| JG646329.1 | Tinospora cordifolia | 840 | 8 Details |
|
| JG647691.1 | Tinospora cordifolia | 824 | 7 Details |
|
| JG650991.1 | Tinospora cordifolia | 865 | 4 Details |
|
| JG650609.1 | Tinospora cordifolia | 742 | 4 Details |
|
| JG647040.1 | Tinospora cordifolia | 843 | 5 Details |
|
| JG649276.1 | Tinospora cordifolia | 754 | 6 Details |
|
| JG650449.1 | Tinospora cordifolia | 764 | 7 Details |
|
| JG645846.1 | Tinospora cordifolia | 729 | 8 Details |
|
| JG650620.1 | Tinospora cordifolia | 747 | 5 Details |
|
| JG650553.1 | Tinospora cordifolia | 810 | 6 Details |
|
| JG650774.1 | Tinospora cordifolia | 846 | 7 Details |
|
| JG648046.1 | Tinospora cordifolia | 817 | 5 Details |
|
| JG650366.1 | Tinospora cordifolia | 768 | 7 Details |
|
| JG650106.1 | Tinospora cordifolia | 787 | 7 Details |
|
| JG647670.1 | Tinospora cordifolia | 739 | 5 Details |
|
| JG645910.1 | Tinospora cordifolia | 872 | 8 Details |
|
| JG646153.1 | Tinospora cordifolia | 875 | 8 Details |
|
| JG650747.1 | Tinospora cordifolia | 839 | 9 Details |
|
| JG646316.1 | Tinospora cordifolia | 787 | 7 Details |
|
| JG649625.1 | Tinospora cordifolia | 805 | 6 Details |
|
| JG648108.1 | Tinospora cordifolia | 807 | 6 Details |
|
| JG646806.1 | Tinospora cordifolia | 615 | 5 Details |
|
| JG646994.1 | Tinospora cordifolia | 637 | 5 Details |
|
| JG648842.1 | Tinospora cordifolia | 774 | 8 Details |
|
| JG648249.1 | Tinospora cordifolia | 607 | 4 Details |
|
| JG648578.1 | Tinospora cordifolia | 766 | 6 Details |
|
| JG647609.1 | Tinospora cordifolia | 852 | 8 Details |
|
| JG646557.1 | Tinospora cordifolia | 618 | 6 Details |
|
| JG647527.1 | Tinospora cordifolia | 811 | 6 Details |
|
| JG647560.1 | Tinospora cordifolia | 713 | 4 Details |
|
| JG648873.1 | Tinospora cordifolia | 829 | 6 Details |
|
| JG647513.1 | Tinospora cordifolia | 739 | 6 Details |
|
| JG646269.1 | Tinospora cordifolia | 669 | 6 Details |
|
| JG648152.1 | Tinospora cordifolia | 779 | 4 Details |
|
| JG650138.1 | Tinospora cordifolia | 778 | 5 Details |
|
| JG648649.1 | Tinospora cordifolia | 842 | 8 Details |
|
| JG645548.1 | Tinospora cordifolia | 799 | 6 Details |
|
| JG646411.1 | Tinospora cordifolia | 786 | 6 Details |
|
| JG646137.1 | Tinospora cordifolia | 797 | 5 Details |
|
| JG647868.1 | Tinospora cordifolia | 832 | 5 Details |
|
| JG646556.1 | Tinospora cordifolia | 820 | 8 Details |
|
| JG647762.1 | Tinospora cordifolia | 802 | 8 Details |
|
| JG647709.1 | Tinospora cordifolia | 725 | 8 Details |
|
| JG650393.1 | Tinospora cordifolia | 837 | 6 Details |
|
| JG648467.1 | Tinospora cordifolia | 837 | 5 Details |
|
| JG649134.1 | Tinospora cordifolia | 531 | 5 Details |
|
| JG650082.1 | Tinospora cordifolia | 735 | 7 Details |
|
| JG645654.1 | Tinospora cordifolia | 843 | 8 Details |
|
| JG650598.1 | Tinospora cordifolia | 794 | 7 Details |
|
| JG650773.1 | Tinospora cordifolia | 792 | 3 Details |
|
| JG648535.1 | Tinospora cordifolia | 807 | 9 Details |
|
| JG648043.1 | Tinospora cordifolia | 788 | 10 Details |
|
| JG650574.1 | Tinospora cordifolia | 821 | 8 Details |
|
| JG646770.1 | Tinospora cordifolia | 771 | 8 Details |
|
| JG649378.1 | Tinospora cordifolia | 798 | 6 Details |
|
| JG650096.1 | Tinospora cordifolia | 773 | 8 Details |
|
| JG649289.1 | Tinospora cordifolia | 654 | 3 Details |
|
| JG645628.1 | Tinospora cordifolia | 813 | 9 Details |
|
| JG650412.1 | Tinospora cordifolia | 819 | 7 Details |
|
| JG650439.1 | Tinospora cordifolia | 770 | 7 Details |
|
| JG647087.1 | Tinospora cordifolia | 819 | 7 Details |
|
| JG648704.1 | Tinospora cordifolia | 757 | 5 Details |
|
| JG646993.1 | Tinospora cordifolia | 718 | 6 Details |
|
| JG650806.1 | Tinospora cordifolia | 847 | 5 Details |
|
| JG650015.1 | Tinospora cordifolia | 817 | 7 Details |
|
| JG648083.1 | Tinospora cordifolia | 823 | 6 Details |
|
| JG649123.1 | Tinospora cordifolia | 800 | 8 Details |
|
| JG647707.1 | Tinospora cordifolia | 873 | 6 Details |
|
| JG645928.1 | Tinospora cordifolia | 799 | 6 Details |
|
| JG646248.1 | Tinospora cordifolia | 187 | 1 Details |
|
| JG648973.1 | Tinospora cordifolia | 621 | 4 Details |
|
| JG650335.1 | Tinospora cordifolia | 787 | 8 Details |
|
| JG649637.1 | Tinospora cordifolia | 835 | 8 Details |
|
| JG647529.1 | Tinospora cordifolia | 691 | 6 Details |
|
| JG650615.1 | Tinospora cordifolia | 809 | 5 Details |
|
| JG650966.1 | Tinospora cordifolia | 800 | 5 Details |
|
| JG649887.1 | Tinospora cordifolia | 643 | 6 Details |
|
| JG646444.1 | Tinospora cordifolia | 797 | 6 Details |
|
| JG649312.1 | Tinospora cordifolia | 774 | 6 Details |
|
| JG647668.1 | Tinospora cordifolia | 726 | 8 Details |
|
| JG647602.1 | Tinospora cordifolia | 795 | 5 Details |
|
| JG646570.1 | Tinospora cordifolia | 626 | 6 Details |
|
| JG648121.1 | Tinospora cordifolia | 694 | 2 Details |
|
| JG649928.1 | Tinospora cordifolia | 145 | 1 Details |
|
| JG650992.1 | Tinospora cordifolia | 784 | 6 Details |
|
| JG650424.1 | Tinospora cordifolia | 856 | 6 Details |
|
| JG646333.1 | Tinospora cordifolia | 777 | 9 Details |
|
| JG649122.1 | Tinospora cordifolia | 761 | 8 Details |
|
| JG645788.1 | Tinospora cordifolia | 694 | 5 Details |
|
| JG649532.1 | Tinospora cordifolia | 804 | 8 Details |
|
| JG646328.1 | Tinospora cordifolia | 800 | 8 Details |
|
| JG645603.1 | Tinospora cordifolia | 703 | 7 Details |
|
| JG648384.1 | Tinospora cordifolia | 741 | 8 Details |
|
| JG646880.1 | Tinospora cordifolia | 638 | 3 Details |
|
| JG650228.1 | Tinospora cordifolia | 828 | 6 Details |
|
| JG648109.1 | Tinospora cordifolia | 764 | 6 Details |
|
| JG650006.1 | Tinospora cordifolia | 752 | 8 Details |
|
| JG651000.1 | Tinospora cordifolia | 794 | 6 Details |
|
| JG647965.1 | Tinospora cordifolia | 796 | 6 Details |
|
| JG648259.1 | Tinospora cordifolia | 466 | 3 Details |
|
| JG647265.1 | Tinospora cordifolia | 774 | 7 Details |
|
| JG647351.1 | Tinospora cordifolia | 837 | 10 Details |
|
| JG647579.1 | Tinospora cordifolia | 738 | 4 Details |
|
| JG647948.1 | Tinospora cordifolia | 636 | 7 Details |
|
| JG648592.1 | Tinospora cordifolia | 708 | 6 Details |
|
| JG646370.1 | Tinospora cordifolia | 800 | 8 Details |
|
| JG649419.1 | Tinospora cordifolia | 782 | 5 Details |
|
| JG650454.1 | Tinospora cordifolia | 637 | 5 Details |
|
| JG646014.1 | Tinospora cordifolia | 808 | 3 Details |
|
| EU815633.1 | Tinospora cordifolia | 77 | 1 Details |
|
| JG645983.1 | Tinospora cordifolia | 798 | 6 Details |
|
| JG647588.1 | Tinospora cordifolia | 818 | 4 Details |
|
| JG645519.1 | Tinospora cordifolia | 759 | 7 Details |
|
| JG650766.1 | Tinospora cordifolia | 808 | 5 Details |
|
| JG646435.1 | Tinospora cordifolia | 821 | 8 Details |
|
| JG650805.1 | Tinospora cordifolia | 834 | 5 Details |
|
| JG647079.1 | Tinospora cordifolia | 755 | 6 Details |
|
| JG647664.1 | Tinospora cordifolia | 152 | 1 Details |
|
| JG647596.1 | Tinospora cordifolia | 792 | 6 Details |
|
| JG649109.1 | Tinospora cordifolia | 195 | 3 Details |
|
| JG646834.1 | Tinospora cordifolia | 592 | 8 Details |
|
| JG650512.1 | Tinospora cordifolia | 790 | 4 Details |
|
| JG648039.1 | Tinospora cordifolia | 786 | 5 Details |
|
| JG646121.1 | Tinospora cordifolia | 775 | 6 Details |
|
| JG650206.1 | Tinospora cordifolia | 740 | 7 Details |
|
| JG649260.1 | Tinospora cordifolia | 653 | 4 Details |
|
| JG646847.1 | Tinospora cordifolia | 731 | 7 Details |
|
| JG646631.1 | Tinospora cordifolia | 829 | 7 Details |
|
| JG649775.1 | Tinospora cordifolia | 776 | 9 Details |
|
| JG646849.1 | Tinospora cordifolia | 748 | 8 Details |
|
| JG646321.1 | Tinospora cordifolia | 733 | 6 Details |
|
| JG646607.1 | Tinospora cordifolia | 774 | 6 Details |
|
| JG647645.1 | Tinospora cordifolia | 832 | 7 Details |
|
| JG649659.1 | Tinospora cordifolia | 774 | 4 Details |
|
| JG649990.1 | Tinospora cordifolia | 723 | 7 Details |
|
| JG646604.1 | Tinospora cordifolia | 839 | 8 Details |
|
| JG649745.1 | Tinospora cordifolia | 586 | 5 Details |
|
| JG646412.1 | Tinospora cordifolia | 860 | 7 Details |
|
| JG648761.1 | Tinospora cordifolia | 807 | 4 Details |
|
| JG649632.1 | Tinospora cordifolia | 816 | 8 Details |
|
| JG650539.1 | Tinospora cordifolia | 828 | 9 Details |
|
| JG650016.1 | Tinospora cordifolia | 815 | 5 Details |
|
| JG647264.1 | Tinospora cordifolia | 830 | 6 Details |
|
| JG647519.1 | Tinospora cordifolia | 774 | 6 Details |
|
| JG646946.1 | Tinospora cordifolia | 705 | 5 Details |
|
| JG648181.1 | Tinospora cordifolia | 678 | 7 Details |
|
| JG649660.1 | Tinospora cordifolia | 759 | 8 Details |
|
| JG648547.1 | Tinospora cordifolia | 807 | 5 Details |
|
| JG645531.1 | Tinospora cordifolia | 654 | 8 Details |
|
| JG649851.1 | Tinospora cordifolia | 794 | 6 Details |
|
| JG648213.1 | Tinospora cordifolia | 557 | 4 Details |
|
| JG650005.1 | Tinospora cordifolia | 796 | 8 Details |
RNA-Seq Information
| Experiment Acc. | Species Name | SRA Study | SRA Run | Treatment | Development Stage | Tissue | Data Accesss |
|---|---|---|---|---|---|---|---|
| SRX11527442 | Tinospora Cordifolia | SRP329561 | SRR15221490 | ||||
| SRX11527441 | Tinospora Cordifolia | SRP329561 | SRR15221491 | ||||
| SRX235296 | Tinospora Cordifolia | SRP018583 | SRR709424 | ||||
| SRX202768 | Tinospora Cordifolia | SRP017022 | SRR675348 | ||||
| SRX130689 | Tinospora Cordifolia | SRP011812 | SRR446984 |
Other non-coding Sequence Information
* If data is not displayed please access the respective tab